jdaviz: [BUG] Cubeviz: MIRI MRS Collapse Fails
Describe the bug Cannot perform any data manipulation on MIRI MRS data using specutils. The working theory is because WCS-TAB is not fully supported somewhere in the stack, but we are not sure where.
To Reproduce Steps to reproduce the behavior:
- Use data file from MAST: jw01024003001_02101_00001_mirifulong_s3d.fits
- Load it into Cubeviz.
- Create subset in spectrum viewer
- Go to collapse plugin
- Set spectral region to subset
- Press Collapse
- See traceback
// ---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/specutils/manipulation/extract_spectral_region.py:29, in _edge_value_to_pixel(edge_value, spectrum, order, side)
28 if hasattr(spectrum.wcs, "spectral"):
---> 29 index = spectrum.wcs.spectral.world_to_pixel(edge_value)
30 else:
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/astropy/wcs/wcsapi/high_level_api.py:309, in HighLevelWCSMixin.world_to_pixel(self, *world_objects)
307 def world_to_pixel(self, *world_objects):
--> 309 world_values = high_level_objects_to_values(*world_objects, low_level_wcs=self.low_level_wcs)
311 # Finally we convert to pixel coordinates
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/astropy/wcs/wcsapi/high_level_api.py:233, in high_level_objects_to_values(low_level_wcs, *world_objects)
232 if callable(attr):
--> 233 world.append(attr(objects[key]))
234 else:
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/astropy/wcs/wcsapi/fitswcs.py:563, in FITSWCSAPIMixin._get_components_and_classes.<locals>.value_from_spectralcoord(spectralcoord)
562 else:
--> 563 return spectralcoord.with_observer_stationary_relative_to(observer).to_value(**kwargs)
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/astropy/units/decorators.py:304, in QuantityInput.__call__.<locals>.wrapper(*func_args, **func_kwargs)
303 with add_enabled_equivalencies(self.equivalencies):
--> 304 return_ = wrapped_function(*func_args, **func_kwargs)
306 # Return
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/astropy/coordinates/spectral_coordinate.py:583, in SpectralCoord.with_observer_stationary_relative_to(self, frame, velocity, preserve_observer_frame)
582 if self.observer is None or self.target is None:
--> 583 raise ValueError("This method can only be used if both observer "
584 "and target are defined on the SpectralCoord.")
586 # Start off by extracting frame if a SkyCoord was passed in
ValueError: This method can only be used if both observer and target are defined on the SpectralCoord.
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/ipyvue/VueTemplateWidget.py:60, in Events._handle_event(self, _, content, buffers)
58 getattr(self, "vue_" + event)(data, buffers)
59 else:
---> 60 getattr(self, "vue_" + event)(data)
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/jdaviz/configs/default/plugins/collapse/collapse.py:49, in Collapse.vue_collapse(self, *args, **kwargs)
47 with warnings.catch_warnings():
48 warnings.filterwarnings('ignore', message='No observer defined on WCS')
---> 49 spec = spectral_slab(cube, spec_min, spec_max)
50 # Spatial-spatial image only.
51 collapsed_spec = spec.collapse(self.selected_func.lower(), axis=-1).T # Quantity
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/specutils/manipulation/extract_spectral_region.py:233, in spectral_slab(spectrum, lower, upper)
204 """
205 Extract a slab from the input `~specutils.Spectrum1D`
206 defined by the lower and upper bounds defined by the ``region``
(...)
229
230 """
231 region = SpectralRegion(lower, upper)
--> 233 return extract_region(spectrum, region)
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/specutils/manipulation/extract_spectral_region.py:149, in extract_region(spectrum, region, return_single_spectrum)
147 extracted_spectrum = []
148 for subregion in region._subregions:
--> 149 left_index, right_index = _subregion_to_edge_pixels(subregion, spectrum)
151 # If both indices are out of bounds then return an empty spectrum
152 if left_index == right_index:
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/specutils/manipulation/extract_spectral_region.py:83, in _subregion_to_edge_pixels(subregion, spectrum)
79 else:
80 # Convert lower value to spectrum spectral_axis units
81 left_reg_in_spec_unit = left_func(subregion).to(spectral_axis.unit,
82 u.spectral())
---> 83 left_index = _edge_value_to_pixel(left_reg_in_spec_unit, spectrum, order, "left")
85 # Right/upper side of sub region
86 if subregion[1].unit.is_equivalent(u.pix):
File ~/miniconda3/envs/mrsoptimal/lib/python3.8/site-packages/specutils/manipulation/extract_spectral_region.py:41, in _edge_value_to_pixel(edge_value, spectrum, order, side)
38 return index
40 except Exception as e:
---> 41 raise ValueError(f"Bound {edge_value}, could not be converted to pixel index"
42 f" using spectrum's WCS. Exception: {e}")
ValueError: Bound 4.890499866509344e-06 m, could not be converted to pixel index using spectrum's WCS. Exception: This method can only be used if both observer and target are defined on the SpectralCoord.
Expected behavior Collapse executes successfully.
About this issue
- Original URL
- State: closed
- Created 2 years ago
- Comments: 18 (10 by maintainers)
Looks like it, although the PR will probably need a PO review.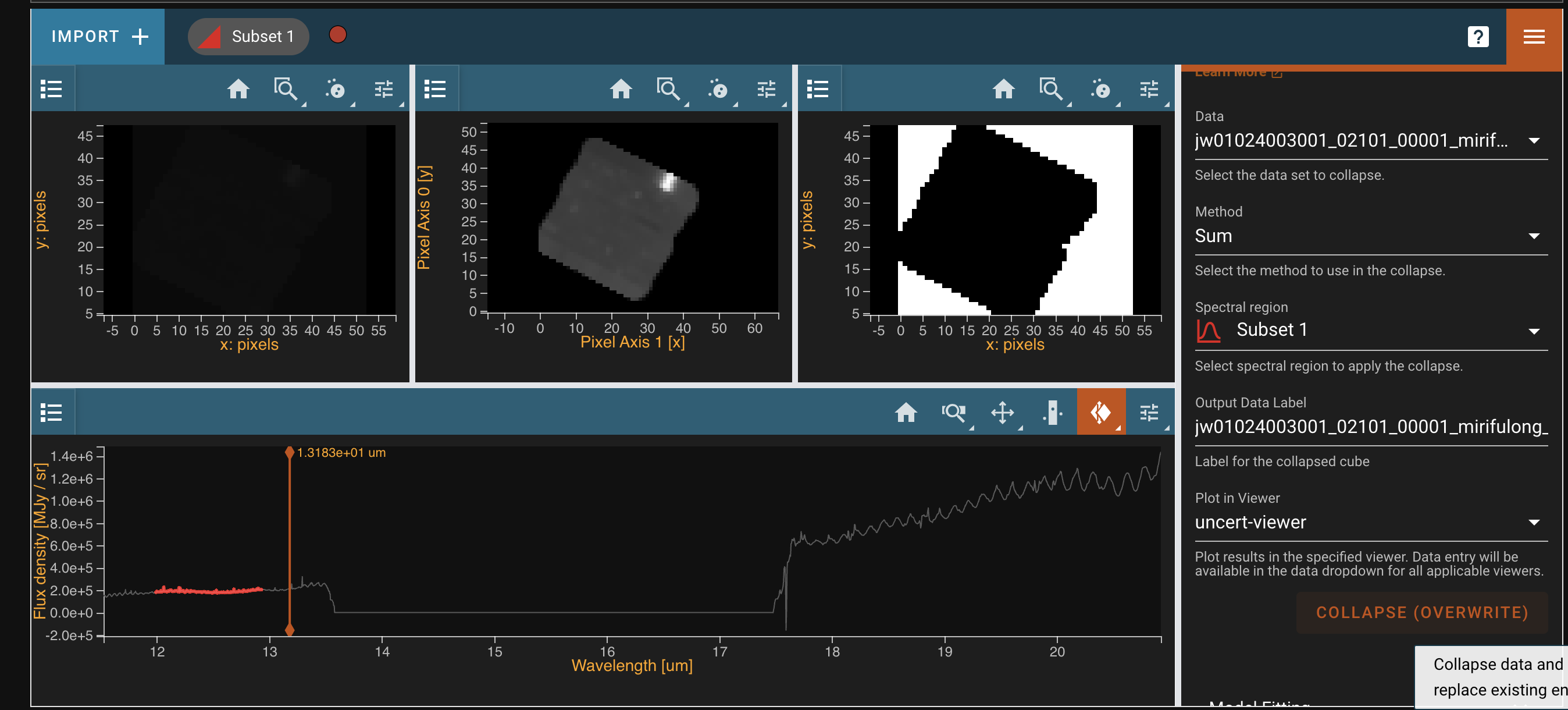
This might be extremely hacky but I got something working with this data
What do you mean “API”? There is no API - just re-arrange the bounding box list.