sktime: [BUG] Handling of numpy2d in transformer Interface differs significantly from time series classification/clustering/regression user expectations
update (@fkiraly’s summary of discussion, point 14-10-2022):
see discussion below. The reported behaviour is not a bug, but a frustrated user expectation.
This stems from how 2D numpy arrays are interpreted in sktime transformers and forecasters, an (m, n) array is interpreted as a single time series with m time points and n variables - let’s call this convention A.
@patrickzib and @TonyBagnall think this is an unnatural way to interpret 2D numpy arrays, and would rather like an (m, n) array to be interpreted as m univariate time series of equal length (number time stamps) n - let’s call this convention B.
This opinion agrees with the way in which @haskarb’s user expectation was frustrated.
Related issue: https://github.com/sktime/sktime/issues/2640 relation is: classifier interpret 2D numpy arrays as per convention B. Everywhere else, convention A is applied. Issue #2640 is about the convention clash between classifiers and everywhere else.
Describe the bug I’m performing MiniRocket tranformation on a small dataset. However there seems to be big difference in runtime. Original repo here.

From Sktime
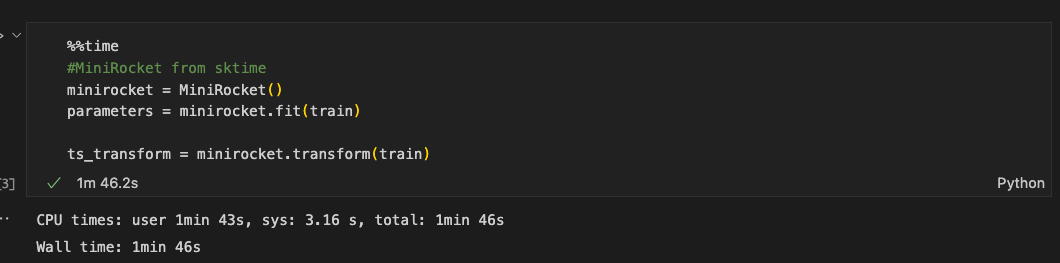
To Reproduce
%%time
#original code from Angus's repo
train = np.random.rand(100,2000).astype('float32')
parameters = minirocket.fit(train)
X_training_transform = minirocket.transform(train, parameters)
%%time
#MiniRocket from sktime
minirocket = MiniRocket()
parameters = minirocket.fit(train)
ts_transform = minirocket.transform(train)
Expected behavior There shouldn’t be so much difference in runtime. As I scale up the number of time-series, and length, this difference will become huge.
Additional context
Versions
About this issue
- Original URL
- State: closed
- Created 2 years ago
- Comments: 34 (14 by maintainers)
Can agree to keep this issue open as a bug?