scanpy: scikit-misc cannot be initiated when sc.pp.highly_variable_genes(adata, n_top_genes=5000, flavor='seurat_v3')
- I have checked that this issue has not already been reported.
- I have confirmed this bug exists on the latest version of scanpy.
- (optional) I have confirmed this bug exists on the master branch of scanpy.
Hello Scanpy, When I’m running sc.pp.highly_variable_genes(adata, n_top_genes=5000, flavor=‘seurat_v3’), it asks me to install scikit-misc, which is already installed. Please see the picture below. Could you please help me to solve this issue? Thanks! Best, YJ Note: Please read this guide detailing how to provide the necessary information for us to reproduce your bug.
Minimal code sample (that we can copy&paste without having any data)
# Your code here
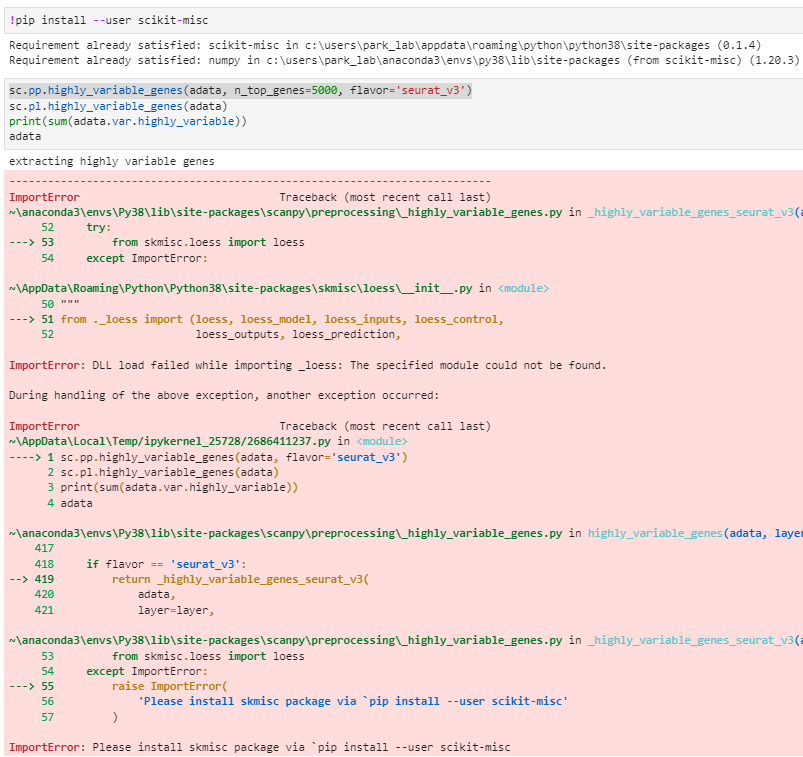
[Paste the error output produced by the above code here]
Versions
3.8
scanpy==1.8.2 anndata==0.7.8 umap==0.5.2 numpy==1.20.3 scipy==1.7.2 pandas==1.3.4 scikit-learn==1.0.1 statsmodels==0.13.1 python-igraph==0.9.8 pynndescent==0.5.5
scvelo==0.2.4 scanpy==1.8.2 anndata==0.7.8 loompy==3.0.6 numpy==1.20.3 scipy==1.7.2 matplotlib==3.5.0 sklearn==1.0.1 pandas==1.3.4
cellrank==1.5.0 scanpy==1.8.2 anndata==0.7.8 numpy==1.20.3 numba==0.54.1 scipy==1.7.2 pandas==1.3.4 pygpcca==1.0.2 scikit-learn==1.0.1 statsmodels==0.13.1 python-igraph==0.9.8 scvelo==0.2.4 pygam==0.8.0 matplotlib==3.5.0 seaborn==0.11.2
[Paste the output of scanpy.logging.print_versions() leaving a blank line after the details tag]
About this issue
- Original URL
- State: closed
- Created 3 years ago
- Comments: 19 (3 by maintainers)
Had this problem, followed the
scikit-miscpackage issue on a related problem and installed the recommended patch withSeems to work now for me.
I found a workaround that does not require downloading the
.whlfile fornumpy=1.19.5. By default, MKL is included when you install numpy with conda. It’s good to do this in a new environment.Now I can run
sc.pp.highly_variable_genes()with no problem.Hello @davidhbrann, Thanks for the suggestion. I found that
numpy‑1.19.5+mkl‑cp36‑cp36m‑win_amd64.whlonly supports python 3.6, but the support of python 3.6 has been dropped by Scanpy. I didn’t findnumpy-1.20.3+mkl-cp38-cp38-win_amd64.whlby google yet, but I believe that your solution will work. It seems thatnumpy‑1.21.5+mkl‑cp38‑cp38‑win_amd64.whlis the only hope, but Scanpy requires numpy<=1.20. I think it’s the compatibility issue. Hope Scanpy can solve this compatibility issue in the future. Thanks! Best, YJI met the same problem one day ago. I run the following: conda install pytorch torchvision torchaudio cudatoolkit=11.3 -c pytorch conda install scvi-tools -c conda-forge conda install -c conda-forge scanpy python-igraph leidenalg It works fine now.
Update: this workaround does not seem to work anymore, at least for scanpy 1.8.2 (you’ll need to
pip install scanpy==1.8.1). Duringpip install scanpy, a newer version of numpy is installed and version 1.19 is overwritten. This newer version does not have MKL, leading us back to square one. It’s also not possible toconda install numpy 1.19as the very last step, because this leads to another error (it’s related to the fact that scanpy needs to be compiled with the same version of numpy).