scipy: Unexpectedly poor results when distribution fitting with `weibull_min` and `exponweib`
My issue is about distribution fitting with weibull_min and exponweib returning clearly incorrect results for shape and scale parameters.
Full details here: https://stats.stackexchange.com/questions/458652/scipy-stats-failing-to-fit-weibull-distribution-unless-location-parameter-is-con
import numpy as np
import pandas as pd
from scipy import stats
x = [4836.6, 823.6, 3131.7, 1343.4, 709.7, 610.6,
3034.2, 1973, 7358.5, 265, 4590.5, 5440.4, 4613.7, 4763.1,
115.3, 5385.1, 6398.1, 8444.6, 2397.1, 3259.7, 307.5, 4607.4,
6523.7, 600.3, 2813.5, 6119.8, 6438.8, 2799.1, 2849.8, 5309.6,
3182.4, 705.5, 5673.3, 2939.9, 2631.8, 5002.1, 1967.3, 2810.4,
2948, 6904.8]
stats.weibull_min.fit(x)
Here are the results:
shape, loc, scale = (0.1102610560437356, 115.29999999999998, 3.428664764594809)
This is clearly a very poor fit to the data. I am aware that by constraining the loc parameter to zero, I can get better results, but why should this be necessary? Shouldn’t the unconstrained fit be more likely to overfit the data than to dramatically under-fit?
And what if I want to estimate the location parameter without constraint - why should that return such unexpected results for the shape and scale parameters?
Scipy/Numpy/Python version information:
import sys, scipy, numpy; print(scipy.__version__, numpy.__version__, sys.version_info)
1.4.1 1.18.1 sys.version_info(major=3, minor=6, micro=10, releaselevel='final', serial=0)
About this issue
- Original URL
- State: closed
- Created 4 years ago
- Comments: 50 (32 by maintainers)
A good start value is crucial for the ML method to find a good solution, especially if many parameters are to be estimated. So if weibul_min had a better _fitstart function like this:
it would solve many situations of unexpectedly poor results when fitting the weibul_min distribution so the user wouldn’t have to fiddle to finetune the fitting. Below I have compared the start values obtained from the default stats.weibull_min._fitstart method the method above. And it is clear that that the bad startvalue for the parameters are the cause of this poor fit:
You mean if providing the guess
loc=xwas a stronger hint, it would be preferable? I see. Even if you provideloc=0, SciPy finds the crazy solution. But perhaps that is because there is not a local minimum of the objective function for it to settle into that is nearloc=0.I’m not sure if we can bake what you’re looking for into the
fitmethod. Maximum Likelihood Estimation is a specific way of fitting a distribution to data, and I think SciPy is doing a reasonable job of that here. You may want to define your own objective function that includes a mathematical description of sanity : )I’m only partially joking. It’s really not too tough to fit using
minimizedirectly. Assuming you’ve already run the code above:produces
This is not much better, but the point is that now you can change the objective function or add constraints to get something closer to what you’re looking for. (It’s just not maximum likelihood estimation anymore.)
(Note: this comment restates and adds some details to some of what @josef-pkt has already said.)
The problem that
fitattempts to solve is to find the maximum of the likelihood function. For theweibull_mindistribution with three parameters, this iswhere
c > 0is the shape parameter, and for brevity,ais the location ands > 0is the scale.Now suppose the x values are sorted in increasing order, so
x[0]is the minimum of all the x values. Also supposecis any value such that0 < c < 1, soc - 1 < 0. Then, asa->x[0]from below, the term((x[0] - a)/s)**(c - 1)blows up, while the other terms in the product approach well-defined limits. So by moving the location parameter closer and closer tox[0], we can make the likelihood function as large as we like. We might say, then, that we should pick the minimum of x as the estimate of the location parameter. But note that we have done that without specifyingcors, other than0 < c < 1ands > 0. So there is not a unique, well-defined solution in this case.The code in SciPy implements
fitby minimizing the negative of the log of the likelihood, and it is attempting to do this using a solver that expects the numerical problem to be well-behaved. But the negative log-likelihood has the same problem as the likelihood: by making the location approach the minimum from below, the negative log-likelihood goes off to negative infinity, for any values of0 < c < 1ands > 0.So we can’t really expect useful results from
weibull_min.fitwhen the location parameter is not fixed. (The same is true forweibull_max, and for some other distributions.) Perhaps the safest choice (already mentioned above) is to simply not allow it, by raising an exception if the location is not fixed.It looks like the
fitfunction is doing what it is supposed to do here, it’s just that the default optimizer is not doing a great job. It would be nice to improve it, but it’s not a simple fix.One thing you could try, @nayefahmad, is to use a different optimizer. For instance, I got pretty good results with
differential_evolutionhere.It’s stochastic, but I’ve gotten as good as:
1.5017832616353088 0.0 3912.5900933784724Although I disagree with the rationale above* for why we can’t expect good results when
locis not defined, the conclusion is correct.As in this post about Nakagami, a plot of the NLLF shows the real reason why either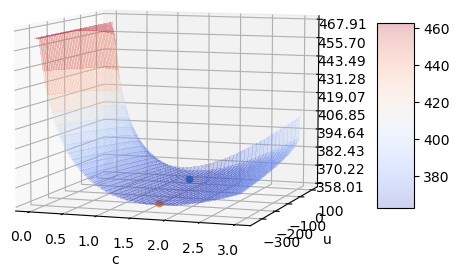
locorcneeds to be fixed - the distribution is nearly over-parameterized forcsufficiently greater than 1. Here is the NLLF as a function ofcandlocwithscaleset at its analytical optimum (for the given values of the other parameters).The blue point is the one
fitgives you whenfloc=0. The orange point is what I got above when I solved the first-order necessary conditions.They’re in a long valley where the NLLF barely changes for large changes in the parameters. The orange point is ever so slightly better in the MLE sense.
(I imagine this is typical for
csufficiently greater than 1. I’ll check some of the other cases above after posting this.)To close this issue, I’d suggest the following:
fitresults, users should fixloc- probably at 0.fitmethod whenlocorcare fixed. In either case, we have an analytical solution for thescale, and we can solve the first-order necessary condition for the remaining parameter withroot_scalar._fitstartsimilar to how we did in Nakagami, but instead of usingloc = np.min(data), make it a little less than that. we can find an approximation forcin the literature, or we can guess using the relationship between the parameters and mean/variance (method of moments, basically. Update: @pbrod gave a suggestion above.fitmethod is frequently having trouble with all parameters free, so if a_fitstartoverride doesn’t help, we should try to override the defaultfitmethod even whenlocandcaren’t fixed. I think that using the analytical solution forscaleinside the objective function will help, or we can use the analytical solution forscaleand try to solve the first-order necessary conditions as above.*For sufficiently small c, the best fit forlocisnp.min(data), whether the LLF is blowing up or not. There is still some sense in which we can maximize the LLF even if we can make it as large as we please. The same thing happens for several distributions; another example of this discussion is in gh-11782, so I won’t repeat it here.Update: @jkmccaughey’s example above has an even more extreme valley.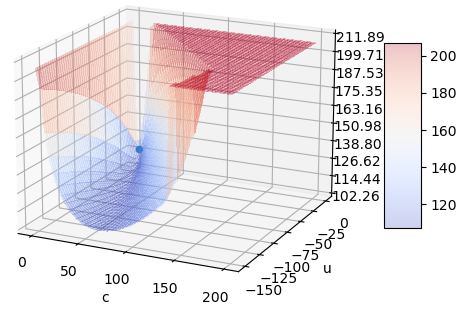
As discussed above, a slightly better MLE than
(6.438181305640674, 0, 21.607076911881855)is(34780967.41119945, -91088178.5479899, 91088200.47560324)!@swallan Maybe this after Nakagami.
Algorithms are weird!
I wrote a little code to investigate:
Qualitatively, both fit the data pretty well: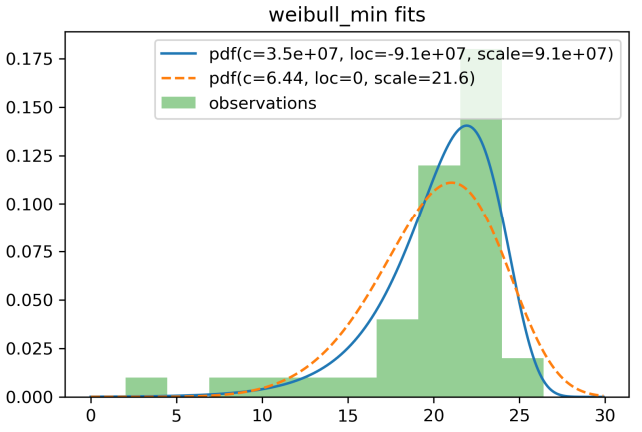
and it turns out that the fit with ridiculous looking numbers (the first one) has a bit higher (less negative) log-likelihood:
It doesn’t sound like you’re doing something incorrectly. You’ve found that for a good estimate, sometimes it helps to fix one of the values. We’re working on improving the starting guesses and adding analytical fits where possible, but if those things are not implemented,
fitjust tries to solve an optimization problem numerically. It doesn’t pay attention to whether the values ofshape,loc, andscalelook reasonable; it just cares about optimizing the objective function (which is the LL function w/ a penalty for data beyond the support of the distribution), and it’s not guaranteed to find the global optimum.weibull_minmay be one of those distributions for which the objective function is relatively flat w.r.t. certain changes of parameters, so providing a good guess or fixing one parameter can go a long way to improving how reasonable the fit parameters are!@nayefahmad You probably don’t need this anymore, but I also get good results by solving the first-order conditions. There is an explicit formula for the scale, and the shape and location can be determined using
root.outputs:
Hi everyone I work in the sector of offshore energy and I analyse data describing meteorological conditions (such as wind speed, wave height, tidal stream speed, …) I also had problems by fitting the weibull_min distribution to data. Through empirically trying different options, I found out that by setting: location parameter first guess = (mean of the data) * 0.3 scale parameter first guess = mean of the data I usually obtain reasonably good fitting. Of course the correct solution is to improve the fitting algorithm for this distribution but, in the meantime, maybe the first guess I’m utilising could be useful for other types of data as well.
OK. There is already an issue gh-11751 that mentions improving the optimizer. I think I’ll combine them. If you’d summarize your takeaways on stackoverflow, I’d appreciate it!
loc=0is a guess for the optimizer to start with. It iteratively improves upon the guess.flocfixes the value oflocto 0.Actually, @nayefahmad , the parameter optimization problem is small enough that
bruteworks quite well.gives
If the objective function in SciPy is correct, then this is a better fit than the one R gave.
Still, I think @nayefahmad has a point - I’ve confirmed that the minimizer is not converging to a great local minimum. I’m going to look into using the other optimizers to see if they work better on this one.
Hi @nayefahmad, I’m working on improving the
fitfunctionality these days (gh-11695, gh-11782), so thanks for submitting this test case. I’ll look into this soon.