scipy: solve is very slow since #6775
Since the merge of #6775 scipy.linalg.solve has been terribly slow for anything but NRHS << N.
This is rather unfortunate because it has left solve unusable (too slow) for anything but NRHS << N. The merge done in #6775 was done primarily based on the performance comparison of gesv and gesvx for NRHS << N. Secondly, in the merge there was no reference of the used LA library, MKL, OpenBLAS, ATLAS, etc.?
Here I show a benchmark on both NRHS << N and NRHS == N which is not every corner case, but should highlight that gesvx is only useful for the former (unless precision is of utmost importance). For the case of NRHS > N the trend follows. I only show performance benchmark for OpenBLAS.
(I also think there is a bug in handling the deprecated flag sym_pos in the solve code. I.e. sym_pos redefines assume_a after its meaning has been digested.)
Reproducing code example:
This code has also been used to reproduce the following images.
from time import time as T
import numpy as np
import scipy as sp
import scipy.linalg as sl
la = sl.lapack
ITT = 25
N = np.arange(100, 1401, 50)
def performance():
t = np.zeros([len(N), 4])
for i, n in enumerate(N):
print(n)
A = np.random.rand(n, n) * 10.
B = np.random.rand(n, 3) * 10.
C = np.random.rand(n, n) * 10.
t0 = T()
for _ in range(ITT):
la.dgesvx(A, B)
t[i, 0] = (T() - t0) / ITT
t0 = T()
for _ in range(ITT):
la.dgesv(A, B)
t[i, 1] = (T() - t0) / ITT
t0 = T()
for _ in range(ITT):
la.dgesvx(A, C)
t[i, 2] = (T() - t0) / ITT
t0 = T()
for _ in range(ITT):
la.dgesv(A, C)
t[i, 3] = (T() - t0) / ITT
def accuracy():
acc = np.zeros([len(N), 4])
for it in range(ITT):
print(it)
for i, n in enumerate(N):
A = np.random.rand(n, n) * 10.
B = np.random.rand(n, 3) * 10.
C = np.random.rand(n, n) * 10.
x = la.dgesvx(A, B)[7]
acc[i, 0] += ((dot(A, x) - B) ** 2).sum() ** .5
x = la.dgesv(A, B)[2]
acc[i, 1] += ((dot(A, x) - B) ** 2).sum() ** .5
x = la.dgesvx(A, C)[7]
acc[i, 2] += ((dot(A, x) - C) ** 2).sum() ** .5
x = la.dgesv(A, C)[2]
acc[i, 3] += ((dot(A, x) - C) ** 2).sum() ** .5
acc /= ITT
performance()
accuracy()
Error message:
There is no error message, only bad performance.
Scipy/Numpy/Python version information:
scipy: 0.19.1
numpy: 1.13.1
OpenBLAS: 0.2.20
Here I show timings T(gesvx) / T(gesv):
NRHS == 3:

NRHS == N:
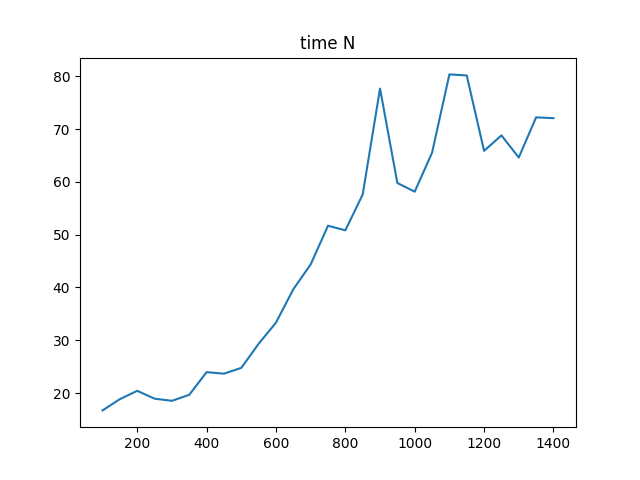
As seen the performance is really bad for NRHS == N. 80 times slower for matrices N >= 1000. I think this justifies a couple of changes to solve:
- Revert default to
gesvroutines, also given the fact thatgesvwas the default routine in previousscipyreleases. - Add an optional flag
refine=Falsewhich determines the use ofgesvxroutines, ifrefine=True. Whether this is an appropriate name for the flag, I don’t know. If you have another idea, let me know.
If you agree to this I would be happy to contribute a PR. I have implemented a copy of solve in one of my own projects to keep using some of the functionality available in scipy, but not in numpy. You may see a re-defined solve here: https://github.com/zerothi/sisl/blob/c04ed44cbaf1ed0d485747045e4bc7cfcf9ecdfa/sisl/_numpy_scipy.py#L86
This also fixes the sym_pos flag. I.e. sym_pos has precedence. If you want a PR and you want assume_a to have precedence, I can do that as well.
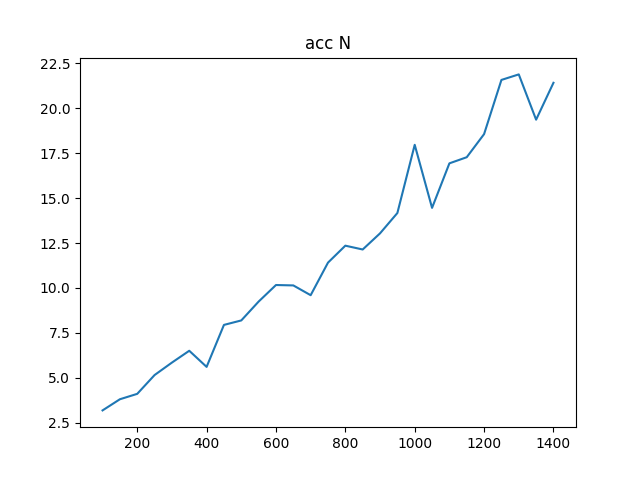
To complete the analysis I have also created an accuracy test to compare the accuracy of gesv and gesvx, they can be seen below where it is clear that gesvx is an order of magnitude more precise (given dot is numerically exact), however the Frobenius norm I would say is showing an adequate precision for this problem.
Yes, this leaves condition numbers and other analysis out of the solve routine. But currently I cannot use solve and I suspect others will also be surprised of the inferior performance since 0.19.1 (or was it 0.19.0?). As you can see in my linked solve I have added an example of how to decide whether refine is applicable for ones application.
The accuracy is shown in two plots, one with the actual Frobenius norm, and one with the relative difference
NRHS == 3:
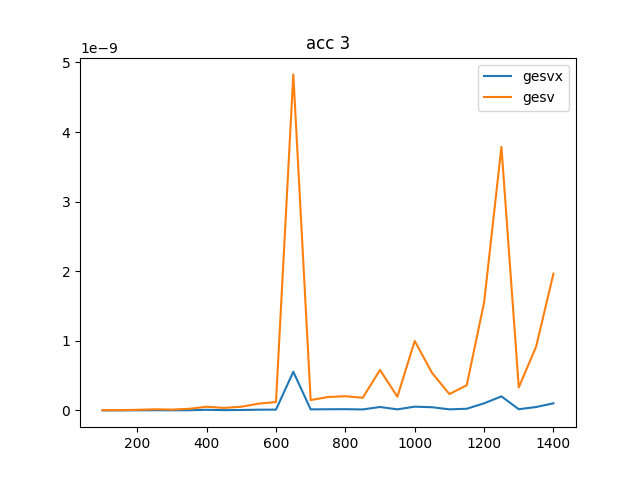
gesv / gesvx
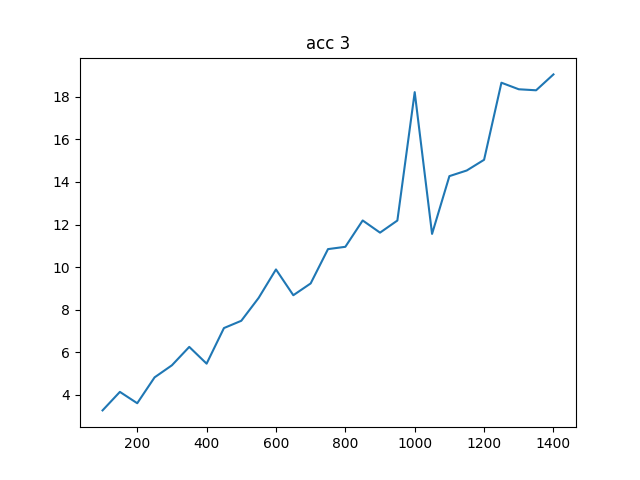
NRHS == N:
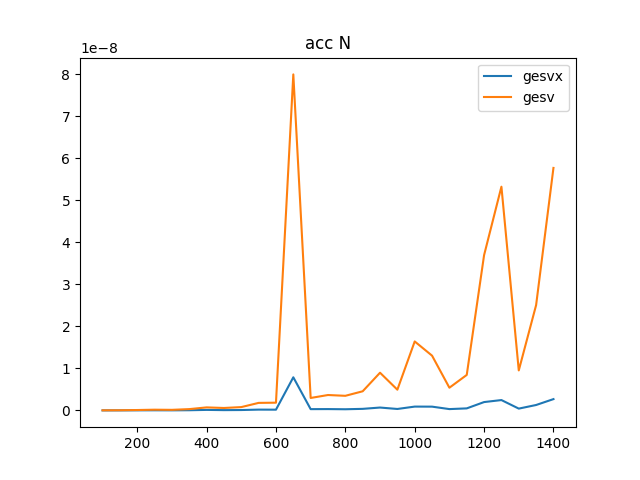
gesv / gesvx
@ilayn, sorry for bashing your PR, your PR is most valuable! I was just surprised when I updated scipy and my code ran 50 times slower!
About this issue
- Original URL
- State: closed
- Created 7 years ago
- Comments: 40 (34 by maintainers)
Commits related to this issue
- ENH: re-enabled old sv lapack routine as defaults This fixes the speed regression in #7847. Since the default change of sv to svx the solve routine suffered a huge performance penalty for anything bu... — committed to zerothi/scipy by zerothi 7 years ago
- ENH: packported #7879 to 1.0.x This fixes the speed regression in #7847. Since the default change of sv to svx the solve routine suffered a huge performance penalty for anything but low order NRHS. ... — committed to zerothi/scipy by zerothi 7 years ago
- BUG: linalg: fix speed regression in solve() This fixes the speed regression in gh-7847. Since the default change of sv to svx the solve routine suffered a huge performance penalty for anything but l... — committed to scipy/scipy by zerothi 7 years ago
- BUG: linalg: fix speed regression in solve() This fixes the speed regression in gh-7847. Since the default change of sv to svx the solve routine suffered a huge performance penalty for anything but l... — committed to rgommers/scipy by zerothi 7 years ago
Sounds great! I have a working solution which I will pr Monday or Tuesday.