rdkit: ERROR SMARTS matching SMILES
Description:
- RDKit Version: 2017.09.3
- Platform: Windows
When matching one SMARTS with a specific SMILES somehow one match is missing.
Following code:
smiles = 'C1=CC=C2C(=C1)C3=CC=CC4=C3C5=C(C=C4)C(C(C=C25)O)O'
mol = Chem.MolFromSmiles(smiles)
q = Chem.MolFromSmarts('[cH]')
mol.GetSubstructMatches(q)
returns:
((0,), (1,), (2,), (5,), (7,), (8,), (9,), (14,), (15,))
It returns 9 hits, although it should find 10. Atom with index 18 is missing. If I draw the image the aromaticity looks correct:
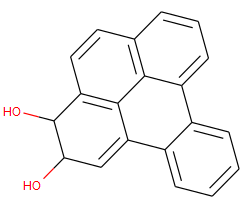
About this issue
- Original URL
- State: open
- Created 6 years ago
- Comments: 27 (18 by maintainers)
SMARTS has no aromaticity model and is very literal. In all your cases where it doesn’t match, you are having aromaticity mismatches. You can see this as follows:
‘O=Cc1ccco1’
Since RDKit doesn’t support both a kekule and aromaticity model simultaneously, this simply isn’t going to match.
I suggest reading this: http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html specifically the smarts vs smiles section.
Cheers, Brian
On Mon, Aug 27, 2018 at 4:39 AM Simon notifications@github.com wrote:
I will say this is one of those corner cases where if you draw the ring as aromatic, it sure looks like there are five bonds to a carbon which makes some of my medicinal chemistry friends look a bit queasy.
Cheers, Brian
On Fri, Aug 24, 2018 at 10:36 AM Brian Kelley fustigator@gmail.com wrote:
This is subtler, the ring is aromatic. RDKit doesn’t store both the aromatic and double bond types, they are independent. The matching patterns are:
pat = Chem.MolFromSmarts(“[#6&H1]:[#6&H1]”)
or
pat = Chem.MolFromSmarts(“[#6&H1]~[#6&H1]”)
~ is any bond, : is aromatic.
Or you could use,
pat = Chem.MolFromSmarts(“[#6&H1]-,=,:[#6&H1]”)
depending.
Cheers, Brian On Fri, Aug 24, 2018 at 10:30 AM Simon notifications@github.com wrote:
The nitrogen is aromatic which is why it isn’t matching.
This is, in general, why using atomic numbers can be better than using symbols. Both the following also match.
pat = Chem.MolFromSmarts(‘[CH3]n’)
pat = Chem.MolFromSmarts(‘[#6&H3][#7]’)
On Fri, Aug 24, 2018 at 8:05 AM Simon notifications@github.com wrote: