pyvista: Trame widget timeout issues on remote server
Describe the bug, what’s wrong, and what you expected.
So I’m trying to run the compute normals example on a headless display. I run Jupyter with jupyter lab --no-browser --port=1234 on the host machine then connect with nohup ssh -NL localhost:1234:localhost:1234 HOSTMACHINE@IP. Before the addition of trame Pyvista did not work with any 3D rendering with this setup (host machine doesn’t have xvfb), but I thought to try it out now with the new backend. I can now run the other examples such as clipping with a surface or surface reconstruction with a 3D interface (yay!), but both examples in the compute normals page result in the connection being terminated. This error occurs before doing the mesh.compute_normals command in the example, but just when plotting the base example mesh. The error also occurs when running the nefertiti example on the same page, so I’m not sure if its an issue with these particular examples or something more sinister. The pyvista window alternates between a blank page (with only the options bar for moving the 3d interface around) to the “connection closed” as shown in the screenshot, alternating after ~1 second. It then repeats the error message below of
WARNING:aiohttp.websocket:Client protocols [‘’] don’t overlap server-known ones () ERROR:root:Exception raised ERROR:root:ConnectionResetError(104, ‘Connection reset by peer’) ERROR:root:Traceback (most recent call last): File “/home/dm575/miniconda3/lib/python3.9/site-packages/wslink/backends/aiohttp/init.py”, line 501, in onMessage await self.sendWrappedMessage( File “/home/dm575/miniconda3/lib/python3.9/site-packages/wslink/backends/aiohttp/init.py”, line 638, in sendWrappedMessage await ws.send_bytes(attachments[key]) File “/home/dm575/miniconda3/lib/python3.9/site-packages/aiohttp/web_ws.py”, line 315, in send_bytes await self._writer.send(data, binary=True, compress=compress) File “/home/dm575/miniconda3/lib/python3.9/site-packages/aiohttp/http_websocket.py”, line 688, in send await self._send_frame(message, WSMsgType.BINARY, compress) File “/home/dm575/miniconda3/lib/python3.9/site-packages/aiohttp/http_websocket.py”, line 659, in _send_frame await self.protocol._drain_helper() File “/home/dm575/miniconda3/lib/python3.9/site-packages/aiohttp/base_protocol.py”, line 87, in _drain_helper await asyncio.shield(waiter) File “/home/dm575/miniconda3/lib/python3.9/asyncio/futures.py”, line 284, in await yield self # This tells Task to wait for completion. File “/home/dm575/miniconda3/lib/python3.9/asyncio/tasks.py”, line 328, in __wakeup future.result() File “/home/dm575/miniconda3/lib/python3.9/asyncio/futures.py”, line 201, in result raise self._exception File “/home/dm575/miniconda3/lib/python3.9/asyncio/selector_events.py”, line 941, in _write_ready n = self._sock.send(self._buffer) ConnectionResetError: [Errno 104] Connection reset by peer
TIA for any help!
Steps to reproduce the bug.
import pyvista as pv
import numpy as np
from pyvista import examples
pv.global_theme.trame.server_proxy_enabled = True
pv.set_jupyter_backend("client")
mesh = examples.download_topo_global()
mesh.plot(cmap="gist_earth", show_scalar_bar=False, notebook=True)
System Information
--------------------------------------------------------------------------------
Date: Wed Feb 08 11:07:34 2023 GMT
OS : Linux
CPU(s) : 72
Machine : x86_64
Architecture : 64bit
RAM : 157.3 GiB
Environment : Jupyter
File system : ext4
GPU Details : error
Python 3.9.15 | packaged by conda-forge | (main, Nov 22 2022, 15:55:03)
[GCC 10.4.0]
pyvista : 0.38.1
vtk : 9.2.5
numpy : 1.24.2
imageio : 2.25.0
scooby : 0.7.1
pooch : v1.6.0
matplotlib : 3.6.3
PyQt5 : 5.15.7
IPython : 8.9.0
ipyvtklink : 0.2.2
scipy : 1.7.1
tqdm : 4.64.1
meshio : 5.3.4
jupyterlab : 3.4.4
pythreejs : 2.3.0
trame : 2.2.6
trame_client : 2.5.1
trame_server : 2.8.1
trame_vtk : 2.0.17
jupyter_server_proxy : 3.2.2
--------------------------------------------------------------------------------
Screenshots
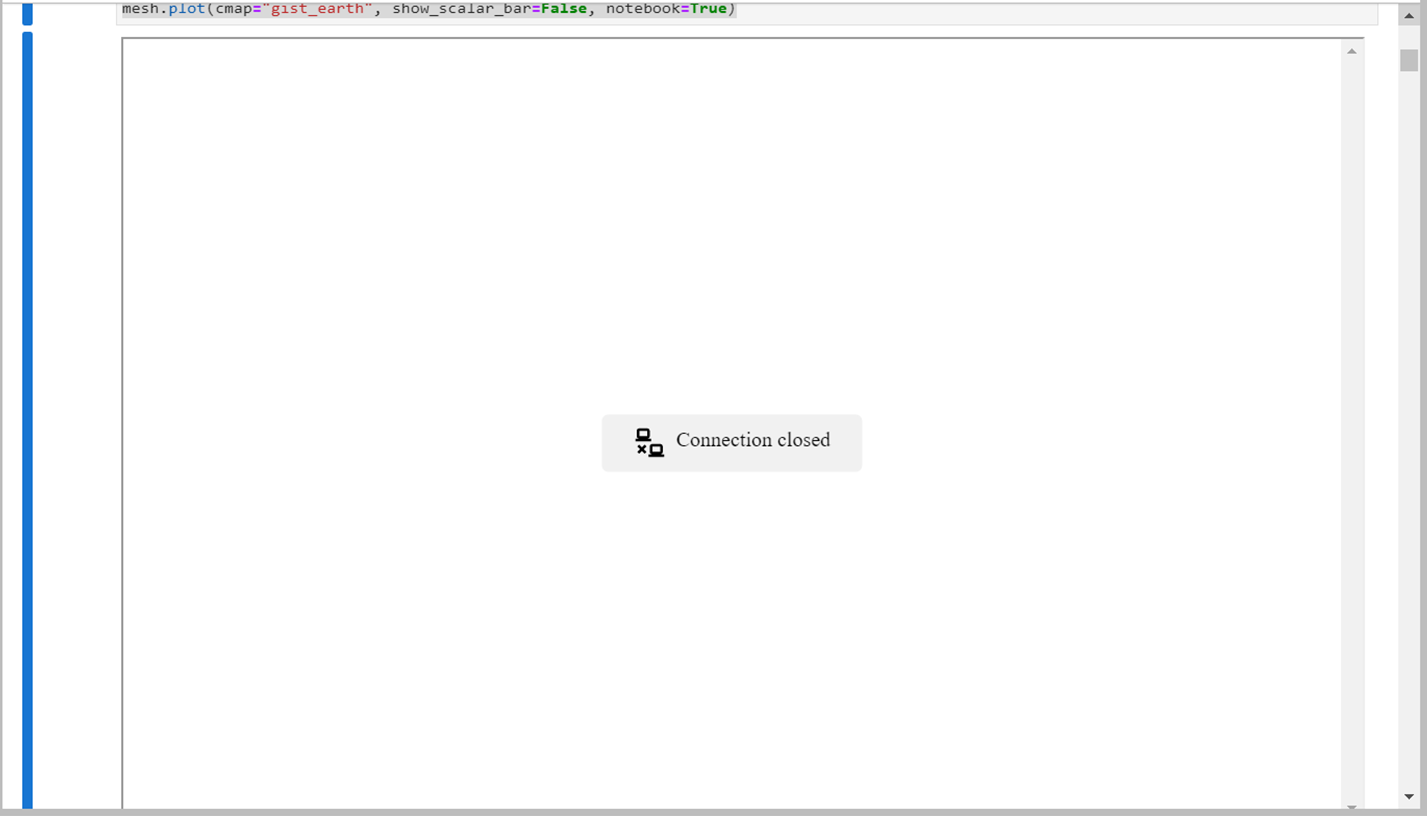
cc @dennissergeev who is also interested in this!
About this issue
- Original URL
- State: closed
- Created a year ago
- Reactions: 3
- Comments: 24 (13 by maintainers)
I’m able to reproduce… I think I know what’s happening and will try to have a patch as soon as I can
I don’t know what happened yesterday – now it is working (able to assign True value)
So I used another server to jump to the host. The command from my PC:
ssh -NL localhost:8892:localhost:8892 USERNAME@FIRSTSERVER.COMThe command from the jump serverssh -NL localhost:8892:localhost:8892 USERNAME@SECONDSERVER.COMThis forwards the port8892which is running Jupyter on the server side. Essentially, I just repeat that but with a different port for the 3D renderer.So this is now resolved - turns out that the amount of data being sent via the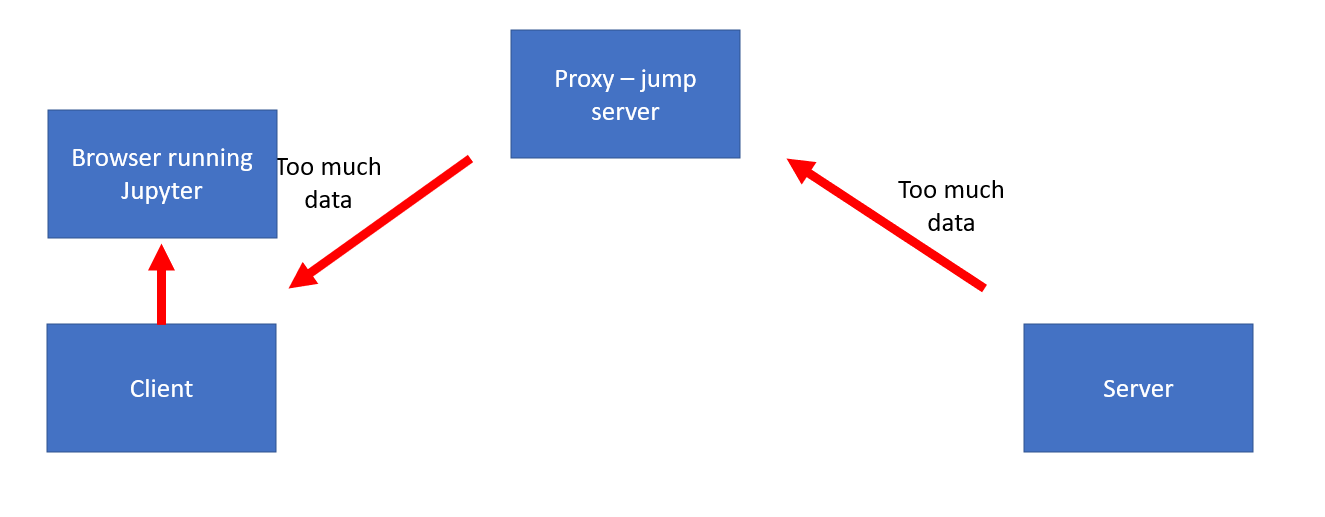 Schematic of the fix:
Schematic of the fix:
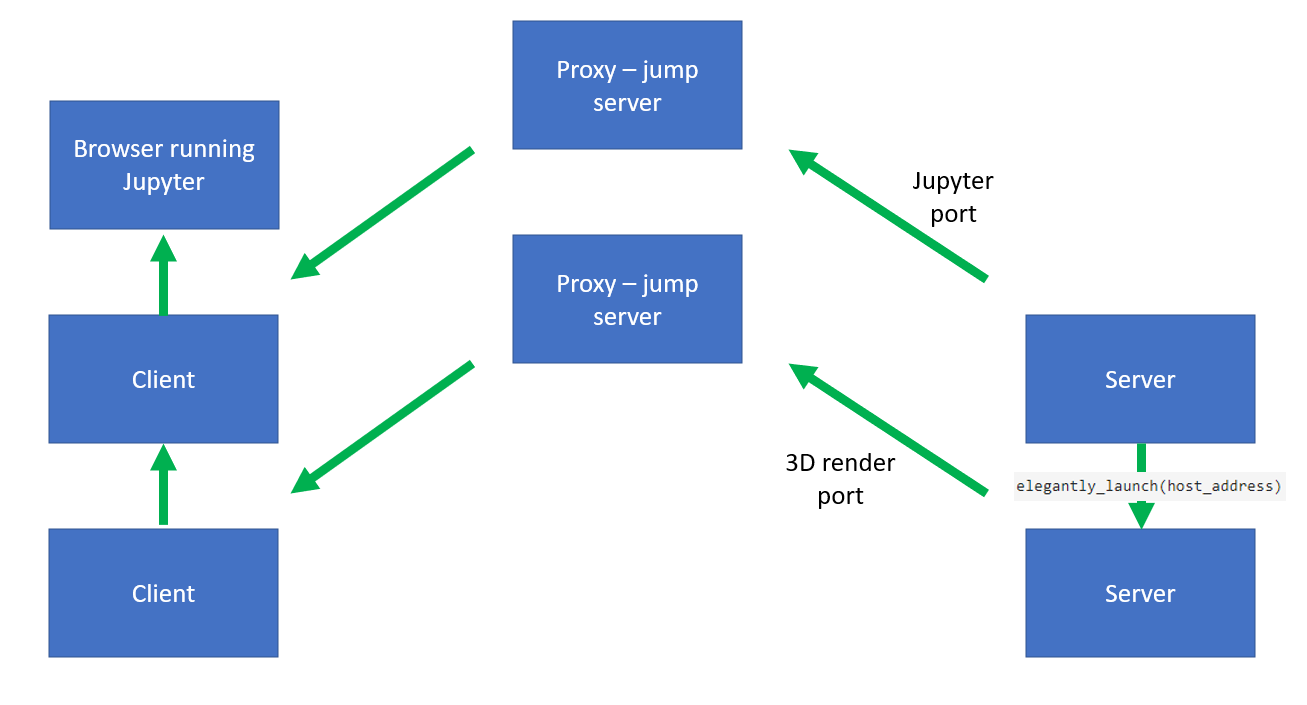
sshand port forwarding was too much for the renderer and the Jupyter lab. The fix was to set the 3D renderer in Jupyter to display from a different port and then port forward to that port as well. Schematic of the problem:Code:
Go with option 2. This is the URL prefix used in your web browser to connect to the remote host.