avogadrolibs: Optimize geometry with Force Field | Error interpreting Open Babel output
Versions:
- Avogadro2: 1.97.0.r837.8ec3eb4-1
- Avogadrolibs: 1.97.0.r2930.d610df7a-1
- Openbabel: openbabel 3.1.1-4
Desktop version: distro: Manjaro Linux base: Arch Linux kernel: 5.10.136-1 arch: x86_64 desktop: GNOME v: 42.4
Description of the bug: I get this error when trying to optimize geometry with a Force Field: “Error interpreting Open Babel output.”
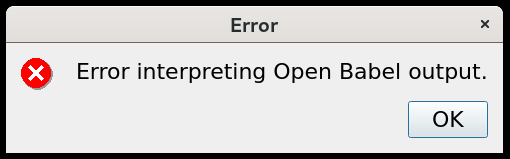
To Reproduce: Steps to reproduce the behaviour:
-
launch
avogadro2 -
Click on “Draw tool”
-
In the left panel menu, “Element”, select “Carbon (6)”, and draw a CH4 molecule.
-
Click on “Extensions” > “Open Babel” > “Configure Force Field” Select “UFF” Force field, “Conjugate Gradient”, “Simple” technique, Select “Energy convergence to 10 ^-6 units” and “Step limit to 100000 steps”
-
Click “Ok”.
-
Click on “Extensions” > “Open Babel” > “Optimize Geometry”
-
The error appears (above screenshot).
Terminal output
1. When launching avogadro2:
$ avogadro2
Using locale: "en_DK"
Extension plugins dynamically found… 40
OBProcess::executeObabel: Running "obabel" "-L formats read"
OBProcess::executeObabel: Running "obabel" "-L formats write"
OBProcess::executeObabel: Running "obabel" "-L forcefields"
OBProcess::executeObabel: Running "obabel" "-L charges"
OBProcess::executeObabel: Running "obabel" "-V"
"obabel" found: "obabel: Open Babel 3.1.0 -- Jul 7 2022 -- 17:58:53"
Checking for "commands" scripts in "/home/david/.local/share/OpenChemistry/Avogadro/commands"
Checking for "commands" scripts in "/home/david/.local/share/flatpak/exports/share/OpenChemistry/Avogadro/commands"
Checking for "commands" scripts in "/var/lib/flatpak/exports/share/OpenChemistry/Avogadro/commands"
Checking for "commands" scripts in "/usr/local/share/OpenChemistry/Avogadro/commands"
Checking for "commands" scripts in "/usr/share/OpenChemistry/Avogadro/commands"
Checking for "commands" scripts in "/var/lib/snapd/desktop/OpenChemistry/Avogadro/commands"
Checking for "commands" scripts in "/usr/bin/../lib/avogadro2/scripts/commands"
Checking for "inputGenerators" scripts in "/home/david/.local/share/OpenChemistry/Avogadro/inputGenerators"
Checking for "inputGenerators" scripts in "/home/david/.local/share/flatpak/exports/share/OpenChemistry/Avogadro/inputGenerators"
Checking for "inputGenerators" scripts in "/var/lib/flatpak/exports/share/OpenChemistry/Avogadro/inputGenerators"
Checking for "inputGenerators" scripts in "/usr/local/share/OpenChemistry/Avogadro/inputGenerators"
Checking for "inputGenerators" scripts in "/usr/share/OpenChemistry/Avogadro/inputGenerators"
Checking for "inputGenerators" scripts in "/var/lib/snapd/desktop/OpenChemistry/Avogadro/inputGenerators"
Checking for "inputGenerators" scripts in "/usr/bin/../lib/avogadro2/scripts/inputGenerators"
Checking for "formatScripts" scripts in "/home/david/.local/share/OpenChemistry/Avogadro/formatScripts"
Checking for "formatScripts" scripts in "/home/david/.local/share/flatpak/exports/share/OpenChemistry/Avogadro/formatScripts"
Checking for "formatScripts" scripts in "/var/lib/flatpak/exports/share/OpenChemistry/Avogadro/formatScripts"
Checking for "formatScripts" scripts in "/usr/local/share/OpenChemistry/Avogadro/formatScripts"
Checking for "formatScripts" scripts in "/usr/share/OpenChemistry/Avogadro/formatScripts"
Checking for "formatScripts" scripts in "/var/lib/snapd/desktop/OpenChemistry/Avogadro/formatScripts"
Checking for "formatScripts" scripts in "/usr/bin/../lib/avogadro2/scripts/formatScripts"
Checking for "charges" scripts in "/home/david/.local/share/OpenChemistry/Avogadro/charges"
Checking for "charges" scripts in "/home/david/.local/share/flatpak/exports/share/OpenChemistry/Avogadro/charges"
Checking for "charges" scripts in "/var/lib/flatpak/exports/share/OpenChemistry/Avogadro/charges"
Checking for "charges" scripts in "/usr/local/share/OpenChemistry/Avogadro/charges"
Checking for "charges" scripts in "/usr/share/OpenChemistry/Avogadro/charges"
Checking for "charges" scripts in "/var/lib/snapd/desktop/OpenChemistry/Avogadro/charges"
Checking for "charges" scripts in "/usr/bin/../lib/avogadro2/scripts/charges"
ScriptLoader::queryProgramName: Unable to retrieve program name for "/usr/lib/avogadro2/scripts/charges/antechamber.py" ; "Error running script '/usr/bin/python /usr/lib/avogadro2/scripts/charges/antechamber.py --display-name --lang en_DK': Abnormal exit status 1 (Unknown error.: Unknown error)\n\nOutput:\nTraceback (most recent call last):\n File \"/usr/lib/avogadro2/scripts/charges/antechamber.py\", line 151, in <module>\n raise RuntimeError(\"antechamber is unavailable\")\nRuntimeError: antechamber is unavailable\n"
ScriptLoader::queryProgramName: Unable to retrieve program name for "/usr/lib/avogadro2/scripts/charges/xtb.py" ; "Error running script '/usr/bin/python /usr/lib/avogadro2/scripts/charges/xtb.py --display-name --lang en_DK': Abnormal exit status 1 (Unknown error.: Unknown error)\n\nOutput:\nTraceback (most recent call last):\n File \"/usr/lib/avogadro2/scripts/charges/xtb.py\", line 100, in <module>\n raise RuntimeError(\"xtb is unavailable\")\nRuntimeError: xtb is unavailable\n"
Open Babel formats ready: 145
b) When performing steps 1-7 above:
OBProcess::executeObabel: Running "obabel" "-icml -ocml --minimize --noh --log --crit 1e-06 --ff UFF --steps 100000 --rvdw 10 --rele 10 --freq 10"
c) When clicking “OK” in the above screenshot:
Open Babel: ""
Process encountered an error, and did not execute correctly.
Exit code: 6
Exit status: QProcess::CrashExit
Exit output: ""
The error message appears again. If I click “OK” again, terminal window prints:
Open Babel: ""
About this issue
- Original URL
- State: closed
- Created 2 years ago
- Comments: 15 (9 by maintainers)
@ghutchis Many thanks for your reply. Please find detailed answers to your questions below.
So,
avogadrolibsandavogadro2were built using Arch Linux AUR repositories, respectively:avogadrolibs-githere: https://aur.archlinux.org/packages?O=0&K=avogadrolibsavogadro2-githere: https://aur.archlinux.org/packages?O=0&K=avogadro2And the way I built those was through the standalone Arch Linux AUR package manager - there are several available, and a very good one is
yay, i.e. simply:yay -S avogadrolibs-gityay -S avogadro2-gitcommands that essentially grab the packages from the links above, and then they do the building automatically.
Regarding Open Babel, I used the
openbabel 3.1.1-4available in Arch Linux, in the section for “Packages”: https://archlinux.org/packages/extra/x86_64/openbabel/ And I installed it following the same command as before:yay -S openbabelSo,
ch4.cifThe terminal window prints:
And the file is generated successfully.
Similarly, if I save as
ch4.mopin, the terminal window prints:And the file is generated successfully.
So,
If I click “OK” I get this:
And the terminal window prints: