fmriprep: Incorrect/dark functional images after SDC with SyN
What happened?
I am testing version 21.0.2 on an fMRI dataset for which fieldmaps were not acquired. As such, I am using the SyN method to attempt susceptibility distortion correction. However, I am getting strange results, inconsistent across subjects. I provide two examples.
- [sub-01] The SDC estimation includes a considerable translation to match the functional and the anatomical images, which results in this estimated fieldmap
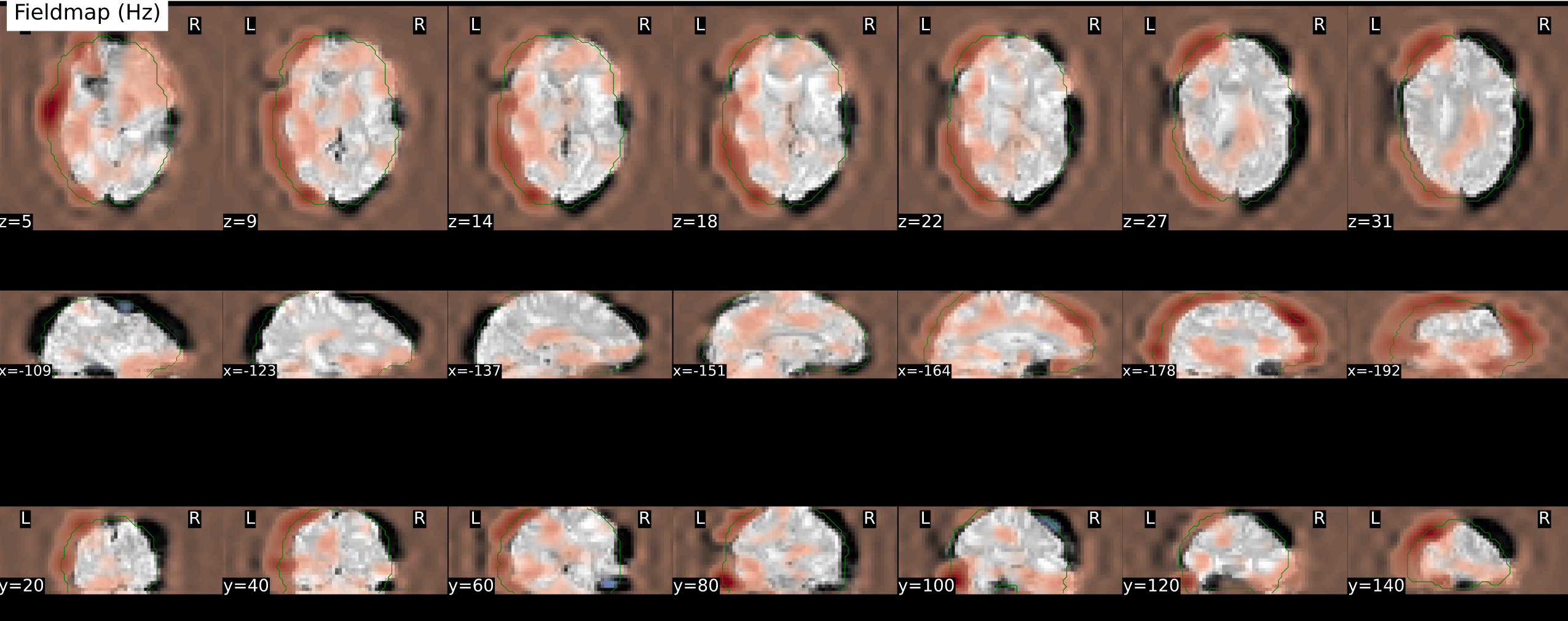
and corrected image:
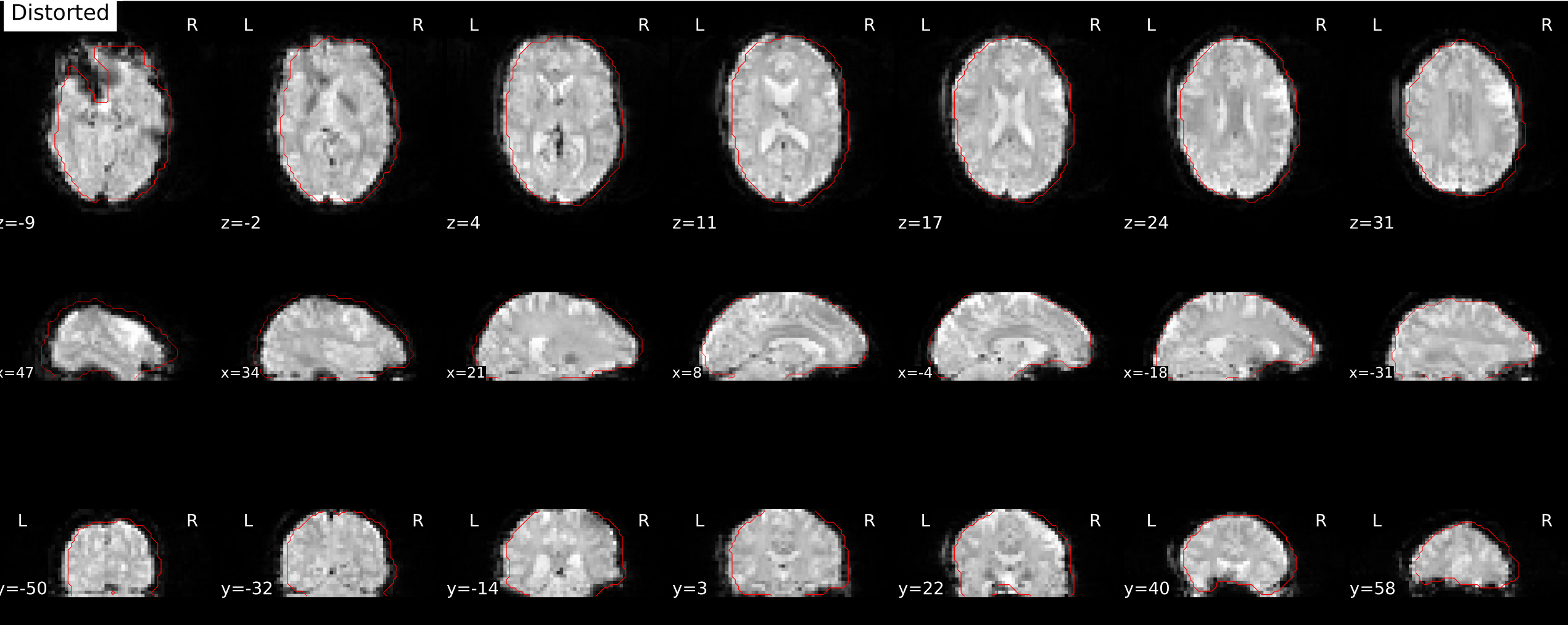
I see no problems in the func to T1w or T1w to MNI registrations.
- [sub-04] The estimated fieldmap seems more reasonable
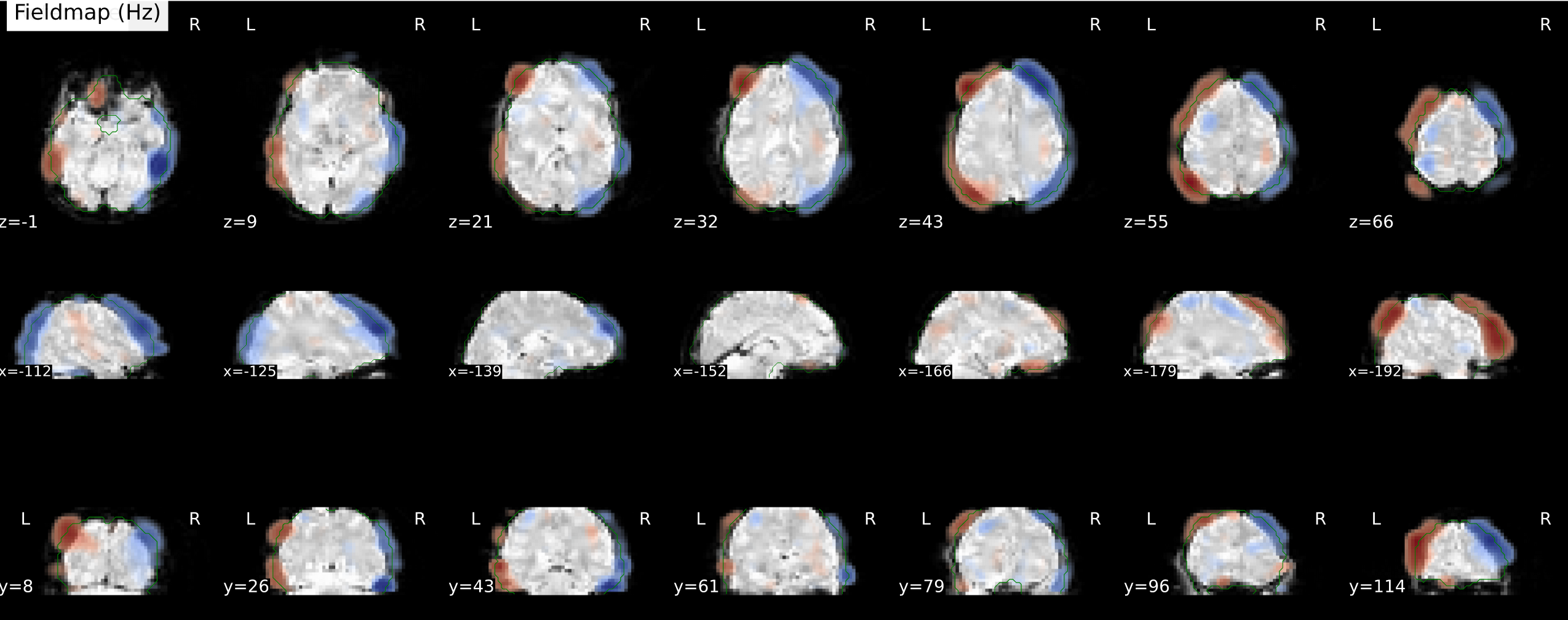
but the corrected image is mostly dark
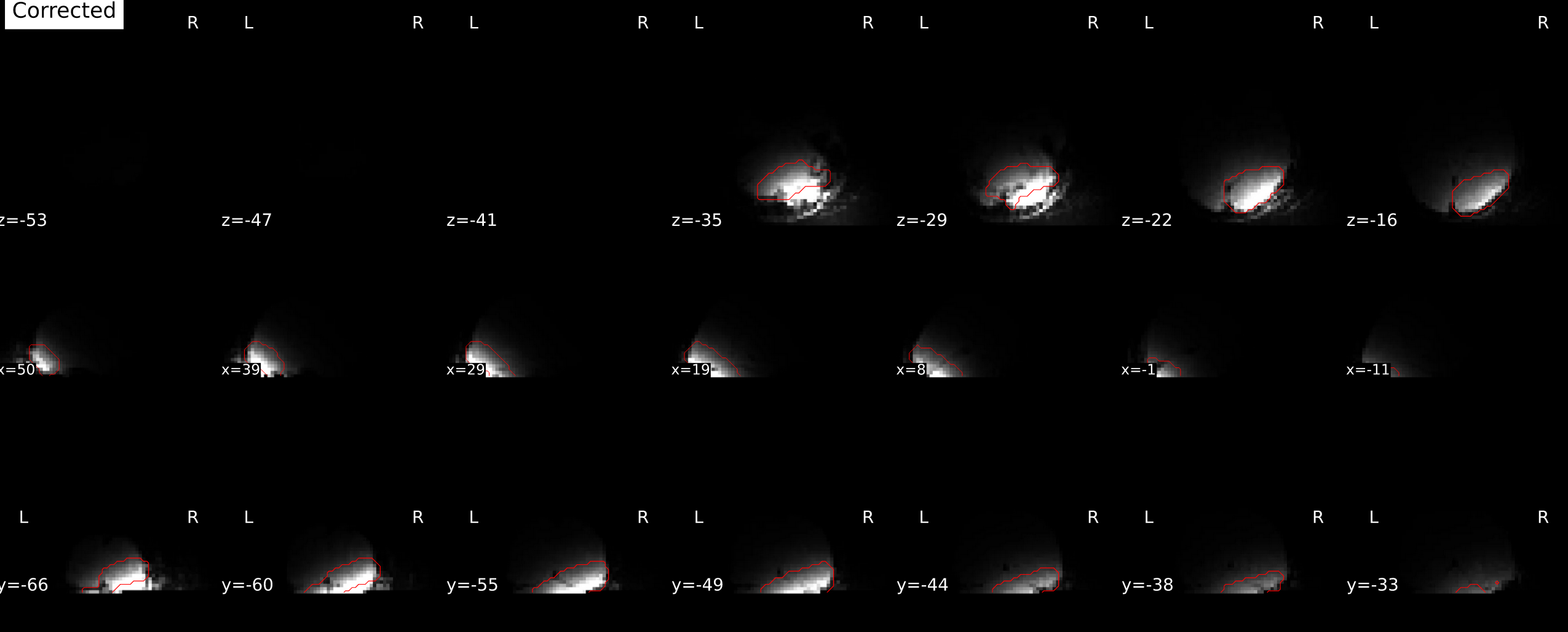
I have tested fmriPrep on the same dataset without the SyN correction and all seems fine.
What command did you use?
docker run -ti --rm \
-v /SCRATCH/users/alexandresayal/BIDS-VP-HYSTERESIS:/data:ro \
-v /SCRATCH/users/alexandresayal/BIDS-VP-HYSTERESIS/derivatives:/out \
-v /SCRATCH/users/alexandresayal/fmriprep-workdir-hyst:/work \
-v /SCRATCH/software/freesurfer/license.txt:/license \
nipreps/fmriprep:21.0.2 \
/data /out/fmriprep \
participant \
-w /work \
--fs-license-file /license \
--use-syn-sdc \
--nprocs 10 \
--mem-mb 35000 \
--stop-on-first-crash \
--fs-no-reconall \
--output-spaces MNI152NLin2009cAsym \
--participant-label 04
What version of fMRIPrep are you running?
21.0.2
How are you running fMRIPrep?
Docker
Is your data BIDS valid?
Yes
Are you reusing any previously computed results?
No
Please copy and paste any relevant log output.
No errors to report in either case.
Additional information / screenshots
I share the .svg images also in this Gdrive folder https://drive.google.com/drive/folders/1RcC4568D6_04gqNGFGp-j4Ec6o8_vdPp?usp=sharing
About this issue
- Original URL
- State: closed
- Created 2 years ago
- Comments: 16 (9 by maintainers)
Please retry with 22.0.2. Skipping SyN-SDC should have been fixed.
Good to hear - the
--use-syn-sdcflag is still fairly experimental, so I can’t say I’m surprised about these discrepancies. Do you know if there is something different (scanning parameters, missing metadata, etc) about the failing participants?If you have any data that you can share with us (T1w + BOLD) of one of the failing runs, it will help us tremendously to improve the robustness of the SyN implementation.
Are there any subjects in the dataset where the SyN correction worked as expected?