napari: Images including NaN pixels cannot be displayed in napari
🐛 Bug
To Reproduce
Steps to reproduce the behavior:
In iPython, you can reproduce this behaviour like so:
%gui qt
import napari
import numpy as np
viewer = napari.Viewer()
data = np.random.random((512, 512)) # make some float type data
float_image = napari.layers.Image(data)
viewer.add_image(data, name='float_data')
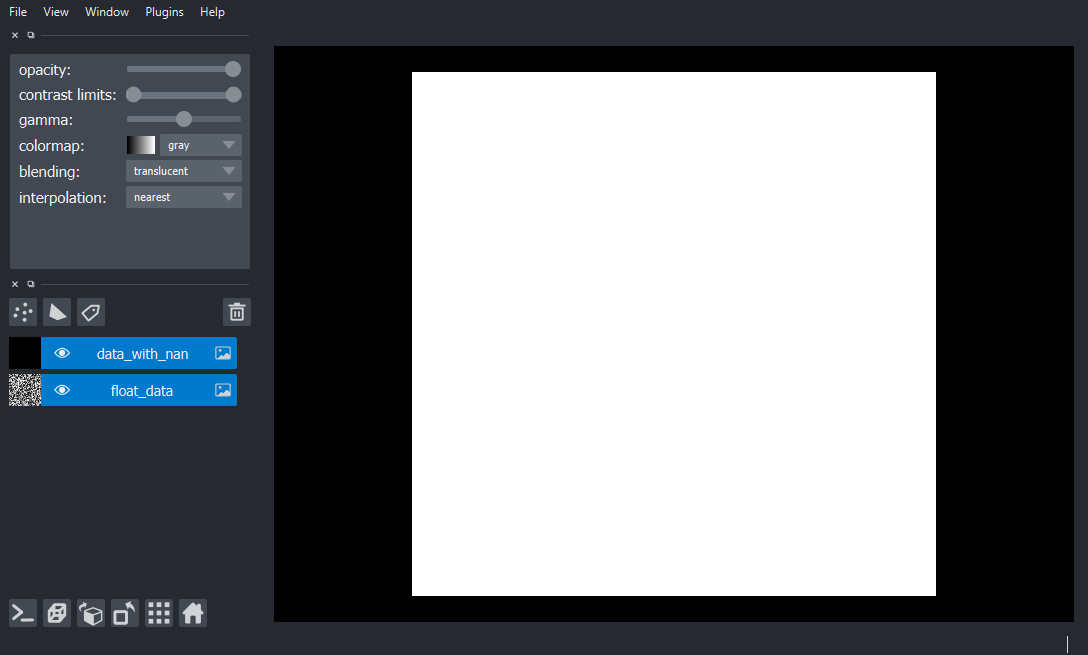
data[0] = np.nan # change a single pixel's value to NaN
viewer.add_image(data, name='data_with_nan') # oh no!
Traceback:
CLICK HERE TO OPEN FULL TRACEBACK
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in less
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in greater
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in less
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in greater
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in less
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in greater
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in less
if color.min() < 0 or color.max() > 1:
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\color\color_array.py:61: RuntimeWarning: invalid value encountered in greater
if color.min() < 0 or color.max() > 1:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-9-b1aa5fd5b5cd> in <module>
----> 1 viewer.add_image(data, name='data_with_nan')
c:\users\genevieve\documents\github\napari\napari\components\add_layers_mixin.py in add_image(self, data, channel_axis, rgb, colormap, contrast_limits, gamma, interpolation, rendering, iso_threshold, attenuation, name, metadata, scale, translate, opacity, blending, visible, multiscale)
228 )
229
--> 230 return self.add_layer(layers.Image(data, **kwargs))
231 else:
232 # Determine if data is a multiscale
c:\users\genevieve\documents\github\napari\napari\components\add_layers_mixin.py in add_layer(self, layer)
60 layer.dims.events.order.connect(self._on_layers_change)
61 layer.dims.events.range.connect(self._on_layers_change)
---> 62 self.layers.append(layer)
63 self._update_layers(layers=[layer])
64
c:\users\genevieve\documents\github\napari\napari\utils\list\_model.py in append(self, obj)
58 def append(self, obj):
59 TypedList.append(self, obj)
---> 60 self.events.added(item=obj, index=len(self) - 1)
61
62 def pop(self, key):
c:\users\genevieve\documents\github\napari\napari\utils\event.py in __call__(self, *args, **kwargs)
506 continue
507
--> 508 self._invoke_callback(cb, event)
509 if event.blocked:
510 break
c:\users\genevieve\documents\github\napari\napari\utils\event.py in _invoke_callback(self, cb, event)
527 self.print_callback_errors,
528 self,
--> 529 cb_event=(cb, event),
530 )
531
c:\users\genevieve\documents\github\napari\napari\utils\event.py in _invoke_callback(self, cb, event)
521 def _invoke_callback(self, cb, event):
522 try:
--> 523 cb(event)
524 except Exception:
525 _handle_exception(
c:\users\genevieve\documents\github\napari\napari\_qt\qt_controls.py in _add(self, event)
65 """
66 layer = event.item
---> 67 controls = create_qt_controls(layer)
68 self.addWidget(controls)
69 self.widgets[layer] = controls
c:\users\genevieve\documents\github\napari\napari\_qt\layers\utils.py in create_qt_controls(layer)
32 Qt controls widget
33 """
---> 34 controls = layer_to_controls[type(layer)](layer)
35
36 return controls
c:\users\genevieve\documents\github\napari\napari\_qt\layers\qt_image_layer.py in __init__(self, layer)
43
44 def __init__(self, layer):
---> 45 super().__init__(layer)
46
47 self.layer.events.interpolation.connect(self._on_interpolation_change)
c:\users\genevieve\documents\github\napari\napari\_qt\layers\qt_image_base_layer.py in __init__(self, layer)
59 self.layer.contrast_limits,
60 self.layer.contrast_limits_range,
---> 61 parent=self,
62 )
63 self.contrastLimitsSlider.mousePressEvent = self._clim_mousepress
c:\users\genevieve\documents\github\napari\napari\_qt\qt_range_slider.py in __init__(self, initial_values, data_range, step_size, collapsible, collapsed, parent)
76
77 self.setRange((0, 100) if data_range is None else data_range)
---> 78 self.setValues((20, 80) if initial_values is None else initial_values)
79 if step_size is None:
80 # pick an appropriate slider step size based on the data range
c:\users\genevieve\documents\github\napari\napari\_qt\qt_range_slider.py in setValues(self, values)
114
115 def setValues(self, values):
--> 116 self.setSliderValues([self._data_to_slider_value(v) for v in values])
117
118 def sliderValues(self):
c:\users\genevieve\documents\github\napari\napari\_qt\qt_range_slider.py in setSliderValues(self, values)
137 self.value_min, self.value_max = values
138 self.valuesChanged.emit(self.values())
--> 139 self.updateDisplayPositions()
140
141 def setStep(self, step):
c:\users\genevieve\documents\github\napari\napari\_qt\qt_range_slider.py in updateDisplayPositions(self)
255 def updateDisplayPositions(self):
256 size = self.rangeSliderSize()
--> 257 range_min = int(size * self.value_min)
258 range_max = int(size * self.value_max)
259 self.display_min = range_min + self.handle_radius
ValueError: cannot convert float NaN to integer
c:\users\genevieve\anaconda3\envs\napari-dev\lib\site-packages\vispy\visuals\image.py:407: RuntimeWarning: invalid value encountered in greater
if clim[1] - clim[0] > 0:
Expected behavior
I expect to be able to see all the not-NaN pixels. In my ideal case NaN pixels would be totally transparent, so you’d never see them no matter what kind of lookup table you use. If that isn’t possible, then I’d be less happy but still ok with rendering NaNs as zeros.
Environment
- Please copy and paste the information at napari info option in help menubar here:
napari: 0.3.1+29.g1b9a8fa
Platform: Windows-10-10.0.17134-SP0
Python: 3.7.7 (default, May 6 2020, 11:45:54) [MSC v.1916 64 bit (AMD64)]
Qt: 5.14.2
PySide2: 5.14.2.1
NumPy: 1.18.4
SciPy: 1.4.1
Dask: 2.16.0
VisPy: 0.6.4
GL version: 4.4.0 - Build 20.19.15.5058
MAX_TEXTURE_SIZE: 16384
Plugins:
- napari-plugin-engine: 0.1.5
- svg: 0.1.2
- Any other relevant information:
Additional context
About this issue
- Original URL
- State: closed
- Created 4 years ago
- Reactions: 3
- Comments: 18 (16 by maintainers)
bumping this. I agree, for scientific work, we definitely need to handle this more gracefully. I think @GenevieveBuckley’s expectation is spot on:
I don’t think so, https://github.com/napari/napari/pull/3701 should have fixed things. I’ll close this, we can always reopen if we discover problems later on.
(TIL there is no walrus emoji. Very sad.)
Note that in skimage we decided against nanmin/nanmaxing everything because there is a significant performance hit, 4-8x depending on your image:
So to compute nanmin/nanmax for a very reasonably sized image you are already at 16ms. The workaround is:
Gets a little more compact in Py3.8+, which we can use after Christmas 😃:
After some quick tests, it appears to be a napari thing. I’ll dive in a bit more!
I’m in favour of waiting and then bumping our vispy requirement, that seem like the simplest thing to do. (Although if anyone wants to make a quick fix in image.py, I’m not opposed to that either)
@scottstanie Lorenzo’s comment https://github.com/napari/napari/issues/1310#issuecomment-937567875 has a few tips you can look at.
I’ve done this here, for the convenience of this discussion: https://github.com/napari/napari/pull/3446
Some good places to start digging into this are:
i think ideally, we’d like to avoid casting/copying data to a safe form if at all possible (that is, a
NaNshould probably stay aNaNinlayer.dataitself). Which kind of means that every place in the code that looks at the data would need to be “ready” for nans. So, things likelayer_utils.calc_data_rangeshould be usingnp.nanmin/np.nanmax, etc… A good general way to test this would be to add some data withNaNto_tests.utils.layer_test_data… which would immediately hit a big surface of the code…