mne-bids: copyfile_eeglab not working
Description
I am currently attempting to copy EEGLAB files via copyfiles_eeglab with a new (BIDS compliant) name. The files are moved and renamed according to the specifications. However, when trying to open those files with MNE there is an error message:
Traceback (most recent call last):
File "<ipython-input-17-8c31a528db9f>", line 1, in <module>
raw_bids = mne.io.read_raw_eeglab('sub-001_ses-D1_task-RS2_run-02_eeg.set')
File "C:\Users\Nils Rosjat\Anaconda3\lib\site-packages\mne\io\eeglab\eeglab.py", line 222, in read_raw_eeglab
eog=eog, verbose=verbose, uint16_codec=uint16_codec)
File "<decorator-gen-216>", line 24, in __init__
File "C:\Users\Nils Rosjat\Anaconda3\lib\site-packages\mne\io\eeglab\eeglab.py", line 326, in __init__
info, eeg_montage, update_ch_names = _get_info(eeg, eog=eog)
File "C:\Users\Nils Rosjat\Anaconda3\lib\site-packages\mne\io\eeglab\eeglab.py", line 145, in _get_info
ch_names, eeg_montage = _get_eeg_montage_information(eeg, has_pos)
File "C:\Users\Nils Rosjat\Anaconda3\lib\site-packages\mne\io\eeglab\eeglab.py", line 123, in _get_eeg_montage_information
ch_pos=dict(zip(ch_names, np.array(pos))),
TypeError: unhashable type: 'numpy.ndarray'
When comparing the original MATLAB structure:
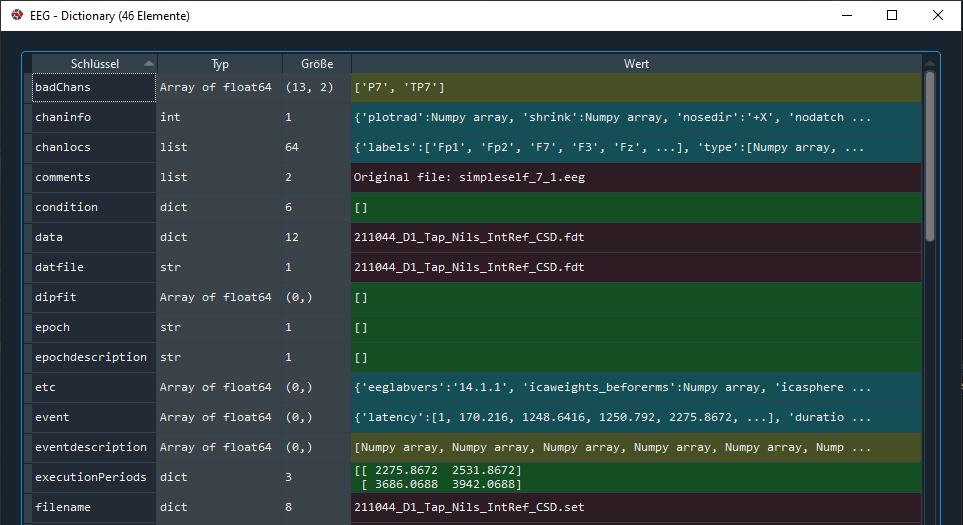
with the newly created structure:
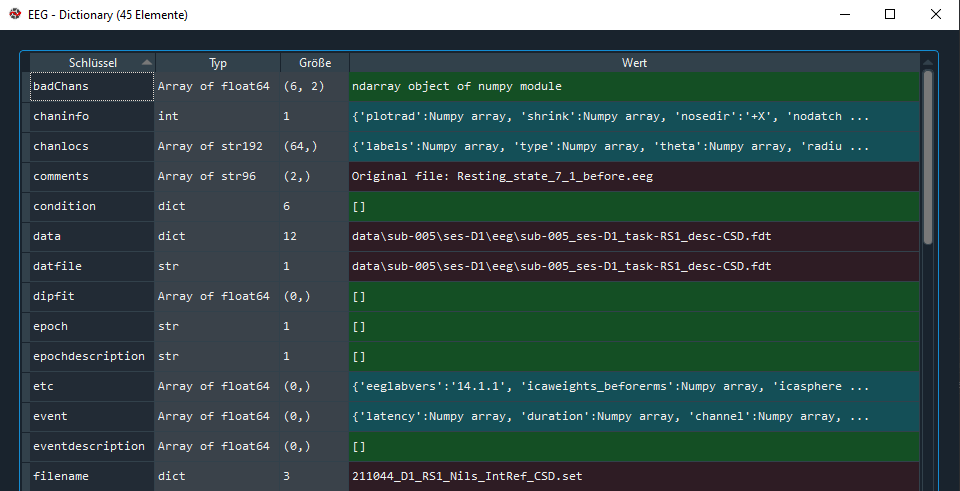
I saw that several entries were replaced by numpy array objects. This seems to happen already in
When replacing loadmat with read_mat (from mne.externals.pymatreader) this seems to work better, but still after saving the dict with savemat the same issue remains.
Script used for conversion
mne_bids version: 0.5
import os.path as op
import os
import mne
from mne_bids import BIDSPath
from mne_bids.copyfiles import copyfile_eeglab
parent_dir = op.abspath('..')
data_path = op.join(parent_dir,'data')
sub_dirs = ['001']
sessions = ['D1']
tasks = ['Tap']
for sub_dir in sub_dirs:
for session in sessions:
for task in tasks:
sub_path = op.join(data_path, sub_dir, 'ses-' + session)
sub_id = os.listdir(sub_path)[0].split('_')[0]
set_name = sub_id + '_' + session + '_' + task + '_Nils_IntRef_CSD.set'
set_file = op.join(data_path, sub_dir, 'ses-' + session,set_name)
set_rename = 'sub-' + sub_dir + '_ses-' + session + '_task-' + task + '_desc-CSD.set'
set_file_renamed = op.join(data_path, 'sub-' + sub_dir, 'ses-' + session, 'eeg', set_rename)
copyfile_eeglab(set_file, set_file_renamed)
About this issue
- Original URL
- State: closed
- Created 4 years ago
- Comments: 25 (19 by maintainers)
Commits related to this issue
- WIP change EEGLAB reading Fixes #652 — committed to hoechenberger/mne-bids by hoechenberger 4 years ago
- [MRG] Make EEGLAB I/O more robust in copyfile_eeglab() (#653) * WIP change EEGLAB reading Fixes #652 * Fix * Reorder imports * Fix I/O roundtrip for example data * Try using loadmat ag... — committed to mne-tools/mne-bids by hoechenberger 4 years ago
Actually, I used that one before and I have no idea why my colleague insisted on using MNE-BIDS for EEGLAB data.
I was also planning to copy and rename some derivates with this function. This has the advantage that I can also add the
_desc-<label>to the filename which is as far as I know not supported withwrite_raw_bids, right?Well… only that nowhere does it enforce BIDS conformity. I’m honestly wondering if these functions currently do more harm than good… (i.e. I don’t think they should be part of the public API unless they ensure they’re actually producing valid BIDS paths)
For now, we just renamed the files with EEGLAB itself.
I’ll try to find a fix for the issue, but any input from @hoechenberger is very appreciated.