pylinac: Apparent regression in BB finding algorithm
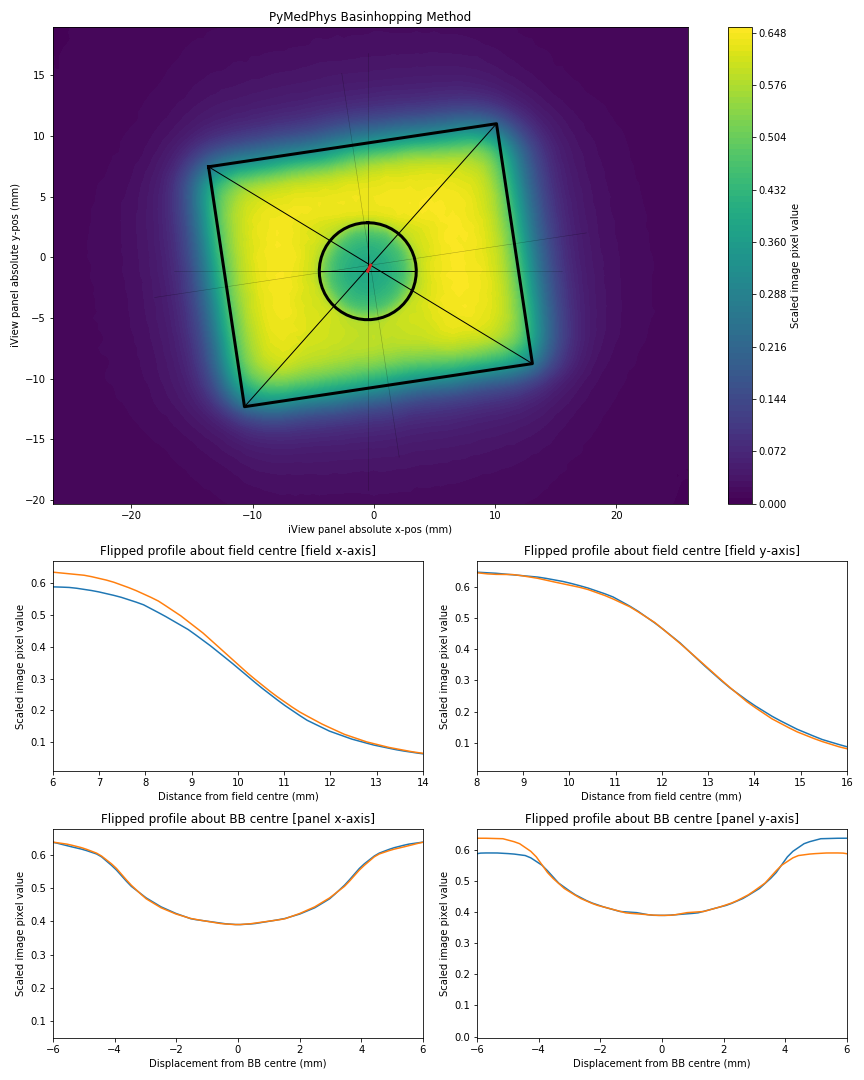
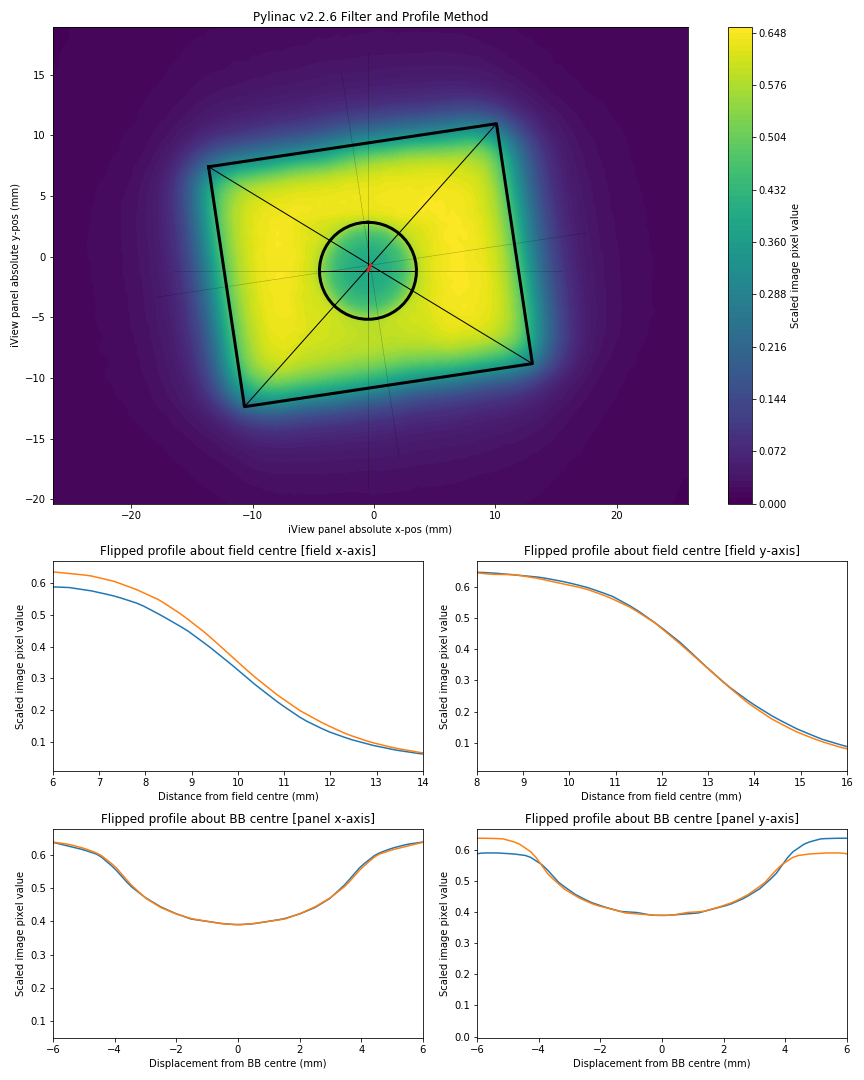
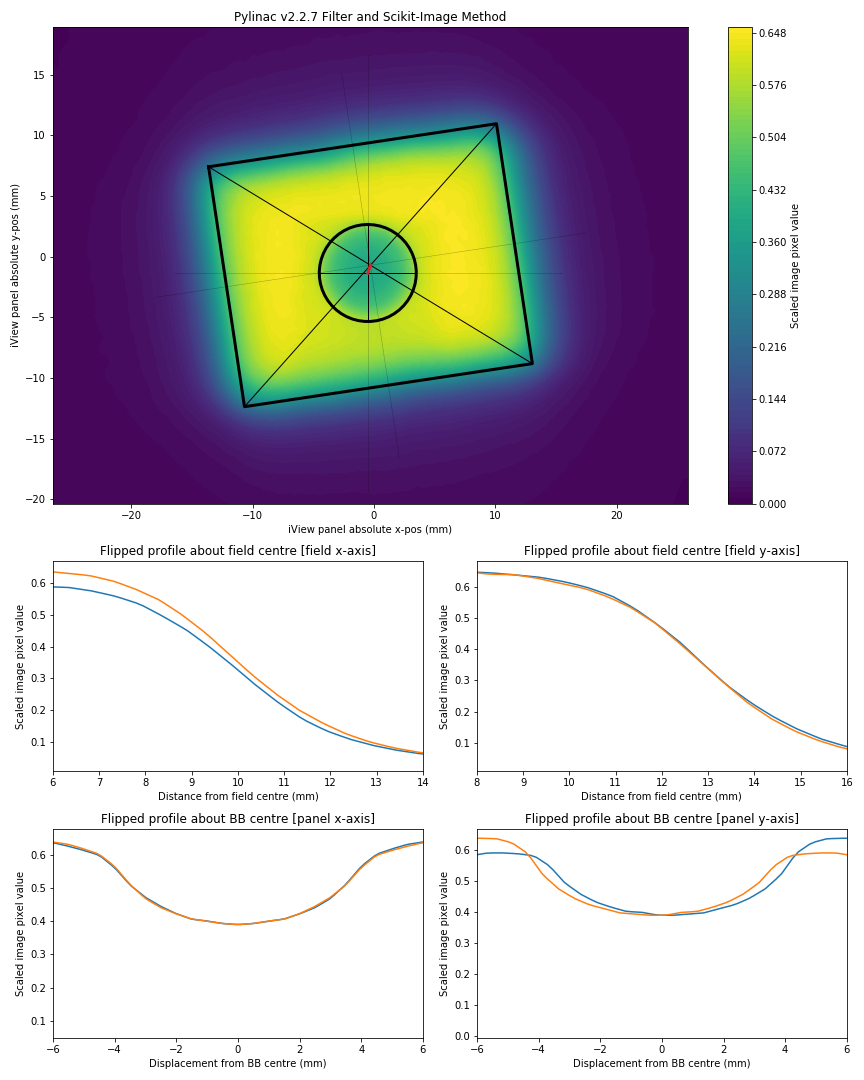
Describe the bug It appears that for some cases, from pylinac version 2.2.6 --> 2.2.7 there has been a regression in the BB finding algorithm. The screenshots given below compare the PyMedPhys wlutz algorithm with the pylinac algorithms from version 2.2.6 and 2.2.7.
Expected behavior Expect a profile taken through the declared centre of the ball bearing should be symmetrical about the BB centre, therefore a flipped profile of the BB in the image should result in overlapping BB penumbras.
Screenshots
To Reproduce
To recreate the above screenshots run the following.
pip install pymedphys==0.17.1
import matplotlib.pyplot as plt
import pymedphys
import pymedphys._wlutz.findfield
import pymedphys._wlutz.findbb
import pymedphys._wlutz.iview
import pymedphys._wlutz.imginterp
import pymedphys._wlutz.reporting
import pymedphys._wlutz.pylinac
image_path = pymedphys.data_path("pylinac_wlutz_regression_image.jpg")
penumbra = 2
edge_lengths = [20, 24]
bb_diameter = 8
x, y, img = pymedphys._wlutz.iview.iview_image_transform(image_path)
field = pymedphys._wlutz.imginterp.create_interpolated_field(x, y, img)
initial_centre = pymedphys._wlutz.findfield._initial_centre(x, y, img)
field_centre, field_rotation = pymedphys._wlutz.findfield.field_centre_and_rotation_refining(
field, edge_lengths, penumbra, initial_centre
)
bb_centre = pymedphys._wlutz.findbb.optimise_bb_centre(
field,
bb_diameter,
edge_lengths,
penumbra,
field_centre,
field_rotation,
pylinac_tol=0.5 # required to ignore diff to pylinac in this case
# pylinac_tol should normally be set to about 0.1 or 0.2
)
pylinac = pymedphys._wlutz.pylinac.run_wlutz(
field, edge_lengths, penumbra, field_centre, field_rotation)
pymedphys._wlutz.reporting.image_analysis_figure(
x,
y,
img,
bb_centre,
field_centre,
field_rotation,
bb_diameter,
edge_lengths,
penumbra,
)
plt.title('PyMedPhys Basinhopping Method')
plt.tight_layout()
plt.savefig('PyMedPhys.png')
pymedphys._wlutz.reporting.image_analysis_figure(
x,
y,
img,
pylinac['v2.2.6']['bb_centre'],
pylinac['v2.2.6']['field_centre'],
field_rotation,
bb_diameter,
edge_lengths,
penumbra,
)
plt.title('Pylinac v2.2.6 Filter and Profile Method')
plt.tight_layout()
plt.savefig('Pylinac v2.2.6.png')
pymedphys._wlutz.reporting.image_analysis_figure(
x,
y,
img,
pylinac['v2.2.7']['bb_centre'],
pylinac['v2.2.7']['field_centre'],
field_rotation,
bb_diameter,
edge_lengths,
penumbra,
)
plt.title('Pylinac v2.2.7 Filter and Scikit-Image Method')
plt.tight_layout()
plt.savefig('Pylinac v2.2.7.png')
Additional context This is using a vendored version of pylinac so that the code can run pylinac 2.2.6 and 2.2.7 at the same time while comparing the results to what PyMedPhys has determined.
Before passing the image to pylinac it is rotated such that the field has no collimator rotation.
About this issue
- Original URL
- State: closed
- Created 5 years ago
- Comments: 20 (3 by maintainers)
Commits related to this issue
- Closes #268 — committed to jrkerns/pylinac by jrkerns 4 years ago
- Merged in feature/RAM-2544_profile_refactor_internal_usage (pull request #268) Profiles part 2: migrate internal Profile usage and improve resampling. Approved-by: Randy Taylor — committed to jrkerns/pylinac by jrkerns 9 months ago
@jrkerns that is some awesome work right there 😃 Thanks James 😃
This is great James. Thank you.
Seconded! Very nice work James!