Stipple.jl: CSVAnalysis does not update `upfiles` after a file is uploaded so the new file is not selectable
The current implementation of the CSVAnalysis demo is not quite right. When a user uploads a new file, the upfiles variable, which is part of Stipple.select(:selected_file; options=:upfiles) and is responsible for listing all available files, is not reactively updated.
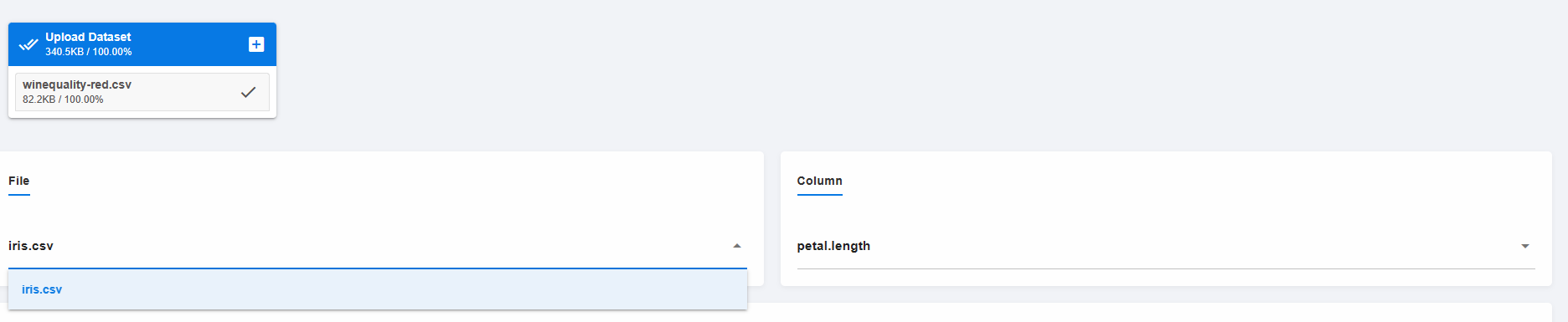
The issue is that onchangeany is not set to trigger when upfiles changes, but only when a selected file or selected column changes. In practice, this means that a user has to first select a different column, which then triggers the handler, which then updates upfiles so that the new file appears (in this demo I made sure to first remove the wineqeuality-red and wineqeuality-white from the uploads directory so simulate a genuine upload of a new file).
@handlers begin
@onchangeany isready, selected_file, selected_column begin
upfiles = readdir(FILE_PATH)
data = CSV.read(joinpath(FILE_PATH, selected_file), DataFrame)
columns = names(data)
datatable = DataTable(data)
if selected_column in names(data)
irisplot = PlotData(x=data[!, selected_column], plot=StipplePlotly.Charts.PLOT_TYPE_HISTOGRAM)
end
end
end
I have a simple fix for this, but I don’t know how to do translate the fix into the new API. The following implementation fixes the issue:
using Stipple
using StippleUI
using StipplePlotly
using CSV
using DataFrames
using Random
const FILE_PATH = "uploads"
mkpath(FILE_PATH)
Genie.config.cors_headers["Access-Control-Allow-Origin"] = "*"
Genie.config.cors_headers["Access-Control-Allow-Headers"] = "Content-Type"
Genie.config.cors_headers["Access-Control-Allow-Methods"] = "GET,POST,PUT,DELETE,OPTIONS"
Genie.config.cors_allowed_origins = ["*"]
@vars AppModel begin
title = "Limited Bandwidth Demo"
selected_file = "iris.csv"
selected_column = "petal.length"
upfiles = readdir(FILE_PATH)
columns = ["petal.length", "petal.width", "sepal.length", "sepal.width", "variety"]
irisplot = PlotData()
files_dirty = false
end
function ui(model)
page(
model,
class = "container",[
row([
cell(class="col-md-12", [
uploader(label="Upload Dataset", accpt=".csv", multiple=true, method="POST", url="http://localhost:8000/", field__name="csv_file")
])
]),
row([
cell(class="st-module", [
h6("File")
Stipple.select(:selected_file; options=:upfiles)
])
cell(class="st-module", [
h6("Column")
Stipple.select(:selected_column; options=:columns)
])
]),
row([
cell(class="st-module", [
h5("Histogram")
plot(:irisplot)
])
])],
)
end
function handlers(model)
Stipple.onany(model.isready, model.selected_file, model.selected_column, model.upfiles) do isready, selected_file, selected_column, upfiles
data = CSV.read(joinpath(FILE_PATH, selected_file), DataFrame)
model.columns[] = names(data)
if selected_column in names(data)
model.irisplot[] = PlotData(x=data[!, selected_column], plot=StipplePlotly.Charts.PLOT_TYPE_HISTOGRAM)
end
end
model
end
model = Stipple.init(AppModel) |> handlers
route("/") do
html(ui(model), context = @__MODULE__)
end
route("/", method = POST) do
@show "Processing upload..."
files = Genie.Requests.filespayload()
for f in files
write(joinpath(FILE_PATH, f[2].name), f[2].data)
end
if length(files) == 0
@info "No file uploaded"
end
# Trigger upfiles change.
model.upfiles[] = readdir(FILE_PATH)
return "Upload finished"
end
up()
Part of the reason it was relatively easy to fix the issue is that I had access to the explicitly defined model, and over time I have been able to piece together a vague mental model of the StippleUI workflow and the fact that the handlers update reactive variables.
I think a drawback of the new UI is that, whilst it tries to abstract away some details and make things more readable, it further obscures the underlying mental model of the StippleUI workflow. If I hadn’t already known that there were Observable variables powering the whole workflow and I first encountered this new API I think I would have struggled even more in fixing my own issues etc.
For instance, the new API has the following route implementation for the CSVAnalysis
route("/", method = POST) do
files = Genie.Requests.filespayload()
for f in files
write(joinpath(FILE_PATH, f[2].name), f[2].data)
end
if length(files) == 0
@info "No file uploaded"
end
upfiles = readdir(FILE_PATH)
return "Upload finished"
end
A newcomer may be perplexed why upfiles is not automatically updated. After all, the upfiles variable was decorated with @out.
@out title = "CSV Analysis"
@out upfiles = readdir(FILE_PATH)
@in selected_file = "iris.csv"
@in selected_column = "petal.length"
@out columns = ["petal.length", "petal.width", "sepal.length", "sepal.width", "variety"]
@out irisplot = PlotData()
I had to play around with @macroexpand to confirm that @handlers is injecting Main_ReactiveModel. in front of variables etc. The expansion shows things like __model__ etc. which I can see are counterparts of model = Stipple.init(AppModel) |> handlers but because all this is hidden behind macros, I have no idea how to access this model variable, let alone ensure that the line
upfiles = readdir(FILE_PATH)
transformed into something like __model__.upfiles = readdir(FILE_PATH).
In summary I have both a question and some feedback. How would one go about introducing my “fix” using the new API? In terms of feedback, I just want to point out that the more work gets done by macros the harder it is to paint a picture of what is really going on, which in turn makes it harder for people to contribute to the codebase etc.
About this issue
- Original URL
- State: closed
- Created a year ago
- Comments: 20 (15 by maintainers)
As promised, the main ingredients are:
Together with a check whether the App looks like
You tested just during we were tagging new versions. StippleUI was not yet released. It does run for me with the latest versions.
Can we close this now?
@zygmuntszpak Can we close this?
If you translate this to the new ReactiveTools API, it would look like this: